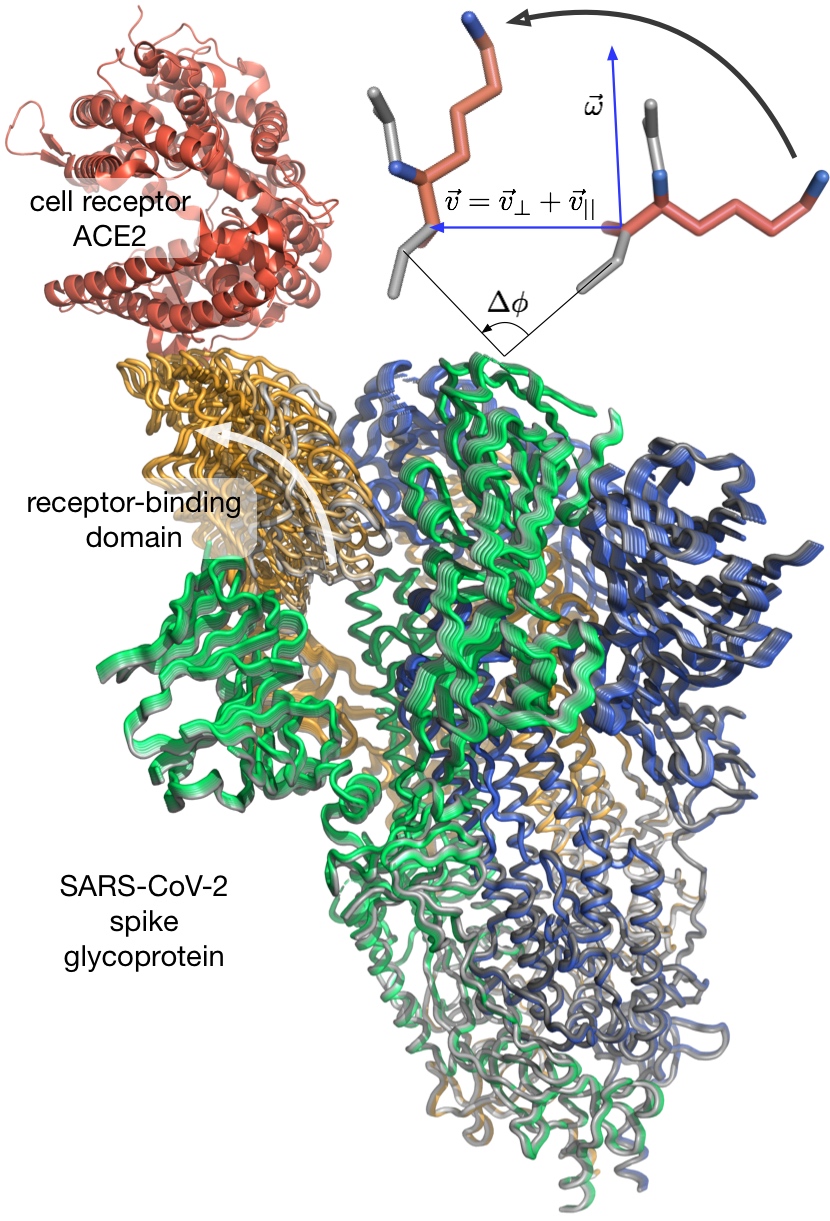
Our work on ‘Predicting protein functional motions: an old recipe with a new twist’ has been just accepted to Biophysical Journal. We report on a real-time method to predict protein functional transitions, using non linear normal mode analysis (NMA). The method extrapolates from instantaneous eigenmotions, computed using the NMA, to a series of twists. We applied it to 155 transitions of different types and showed that it produces better conformations than the linear NMA, and achieves higher transition coverage. In particular, it is able to describe localized motions, although the NMA has been reputed to be unsuitable for this type of motions. Our method is available as a part of the NOn-Linear rigid Block (NOLB) package.