| About | Method | Validation | Download | Usage | References | Citing |
About
VoroCNN is a novel method for local residue-level quality assessment of protein folds. The method is based on a deep convolutional neural network (CNN) constructed on a Voronoi tessellation of 3D molecular structures. The method is developed together with Česlovas Venclovas’ lab at Vilnius University.
Method
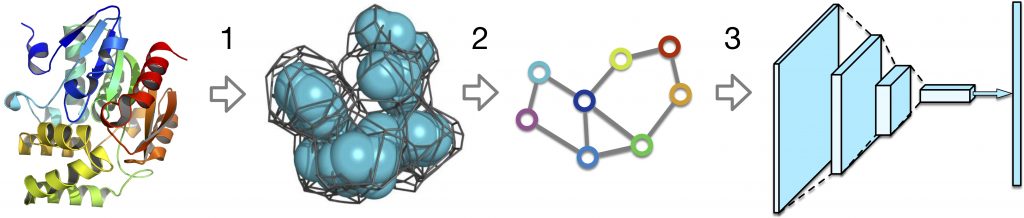
Firstly, a Voronoi tessellation of a 3D-model is computed with Voronota [1]. Then, based on Voronoi 3D-tessellation, a graph is built. Finally, a graph convolutional neural network predicts local CAD-scores for all residues in the initial model.
Validation
We validated the VoroCNN method on CASP11 and CASP12 submissions and also visually inspected the prediction results. For example, the figure below illustrates VoroCNN’s local scores predictions. The colorbar on the right correspond to the values of the scores. A) VoroCNN predictions for the T0951 CASP target (left), T0951TS498_1 model (center), and the ground-truth local CAD-scores (right). B) VoroCNN predictions for the T0954 CASP target (left), T0954TS112_3 model (center), and the ground-truth local CAD-scores (right). C) VoroCNN predictions for the obligatory complex of bacteriophage RNA-binding protein (pdb code 1UNA, left) and its individual subunits (right). D) VoroCNN predictions for the obligatory complex of cyclohexadienyl dehydrogenase (pdb code 4WJI, left) and its individual subunits (right).
Download
The method and the results on test sets are available in the VoroCNN GitLab repository. Please follow the instructions about the installation for Linux and MacOS operating systems.
Usage
VoroCNN uses Voronota by Kliment Olechnovic (kliment@ibt.lt) in order to construct the tessellation and build the graph. Prior to running VoroCNN, Voronota must be installed. You can use already compiled executables for MacOS or Linux. Once Voronota is installed, VoroCNN can be run by executing the precompiled file vorocnn :
Please make sure you have changed the access permissions of vorocnn to make it executable as chmod +x vorocnn.
If you want to process only one model, just specify the path to the model PDB file in argument -i and pass the path of Voronota executable file in argument -v :
|
1 |
./vorocnn -i /path/to/model.pdb -v /path/to/voronota |
If you want to process multiple models, specify the path to the directory with model PDB files in argument -i and pass the path of Voronota executable file in argument -v :
|
1 |
./vorocnn -i /path/to/models/ -v /path/to/voronota |
VoroCNN will create folder vorocnn_output/ with the results in the current directory.
It is recommended to score multiple structures in one run as starting of the executable file vorocnn may take up to 30 seconds.
References
[1] Olechnovič, Kliment, and Česlovas Venclovas. “Voronota: a fast and reliable tool for computing the vertices of the Voronoi diagram of atomic balls.” Journal of computational chemistry 35.8 (2014): 672-681.