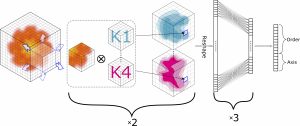
DeepSymmetry is a method based on three-dimensional (3D) convolutional networks that detects structural repetitions in proteins and their density maps. It identifies tandem repeat proteins, proteins with internal symmetries, their symmetry order, and also the corresponding symmetry axes.
DeepSymmetry is available for each platform here.
This archive contains binaries to create density maps from pdb files, the model that we trained to detect symmetries, a few examples to test the model, and a python script that is able to read the model and to apply it to pdb files. It requires TensorFlow to be installed.
Reference:
Guillaume Pagès & Sergei Grudinin, “DeepSymmetry : Using 3D convolutional networks for identification of tandem repeats and internal symmetries in protein structures“. Bioinformatics, Oxford University Press (OUP), In press, ⟨10.1093/bioinformatics/btz454⟩