https://gitlab.inria.fr/grudinin/sbrod
About
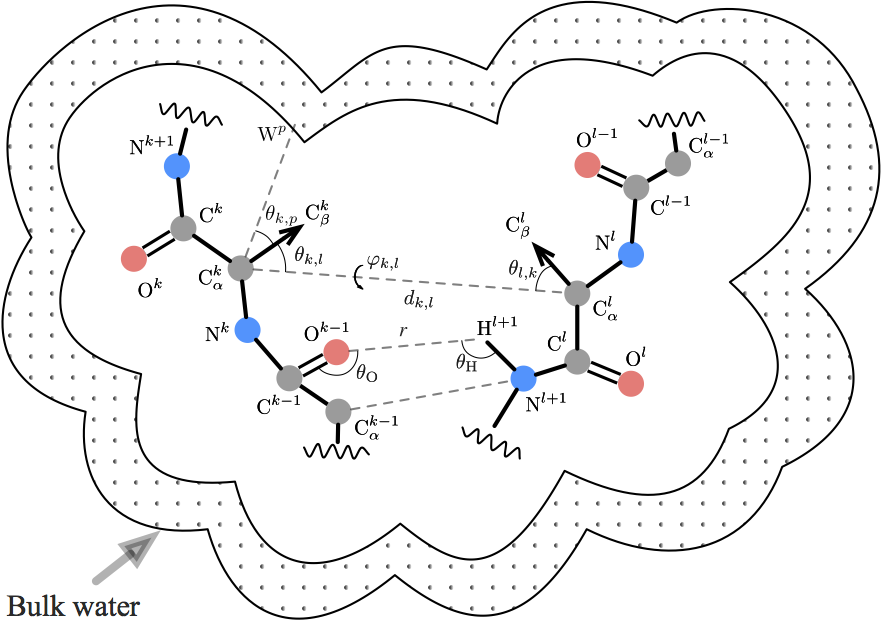
Smooth orientation-dependent scoring function (SBROD) for coarse-grained protein quality assessment uses only the conformation of the protein backbone, and hence it can be applied to scoring the coarse-grained protein models.
Method
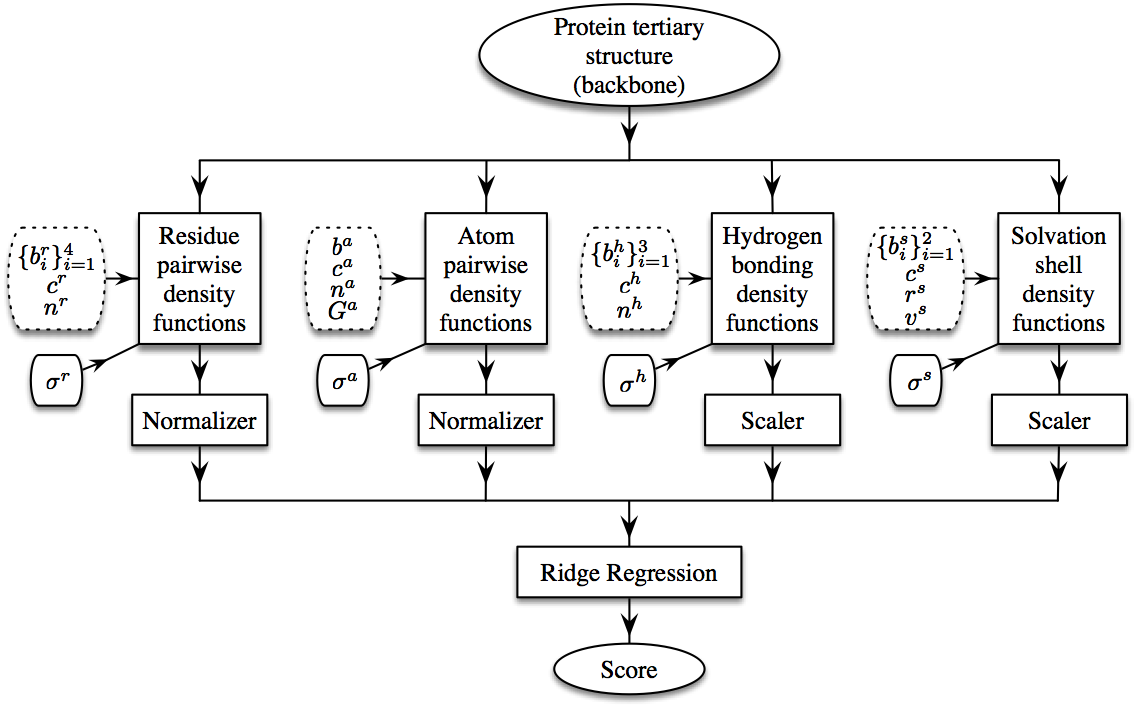
Workflow of the SBROD method. The dotted blocks correspond to adjustable structural parameters that are to be chosen at the training stage.
The workflow of SBROD consists in two stages. First, the method extracts features from each protein model in the dataset. Then, the scoring function assigns a score to each processed protein model depending on its features extracted at the first stage. Figure above schematically shows the workflow of SBROD. Here, four types of inter-atomic interactions, described in details below, are taken into account when extracting the features. After these features have been extracted and preprocessed, a Ridge Regression model is trained on them to predict the GDT-TS of protein models.
Authors
Mikhail Karasikov1-3, Guillaume Pagès3 & Sergei Grudinin3,
1Center for Energy Systems, Skolkovo Institute of Science and Technology, Moscow, 143026, Russia 2Moscow Institute of Physics and Technology, Moscow, 141701, Russia3Nano-D team, Inria/CNRS Grenoble, France
e-mail: Sergei.Grudinin @ inria.frDownload
Binaries, source code and tutorials can be found at https://gitlab.inria.fr/grudinin/sbrod
The server is running at www.karasikov.com/proteins/
SBROD was also evaluated in CASP13 as a QA server.
References
- Mikhail Karasikov, Guillaume Pagès & Sergei Grudinin, “Smooth orientation-dependent scoring function for coarse-grained protein quality assessment”. Bioinformatics, bty1037, https://doi.org/10.1093/bioinformatics/bty1037
License
All rights reserved. The academic version is free.