Software
 |
BioImage-ITBioImageIT is a France-BioImaging project. It aims at providing good IT practices, recipies and tools for imaging facilities to deploy interoperable image processing and data analysis solutions. BioImageIT is also a set of tools to facilitate the installation of image management and analysis facility OS: Windows, Mac OSX, Linux |
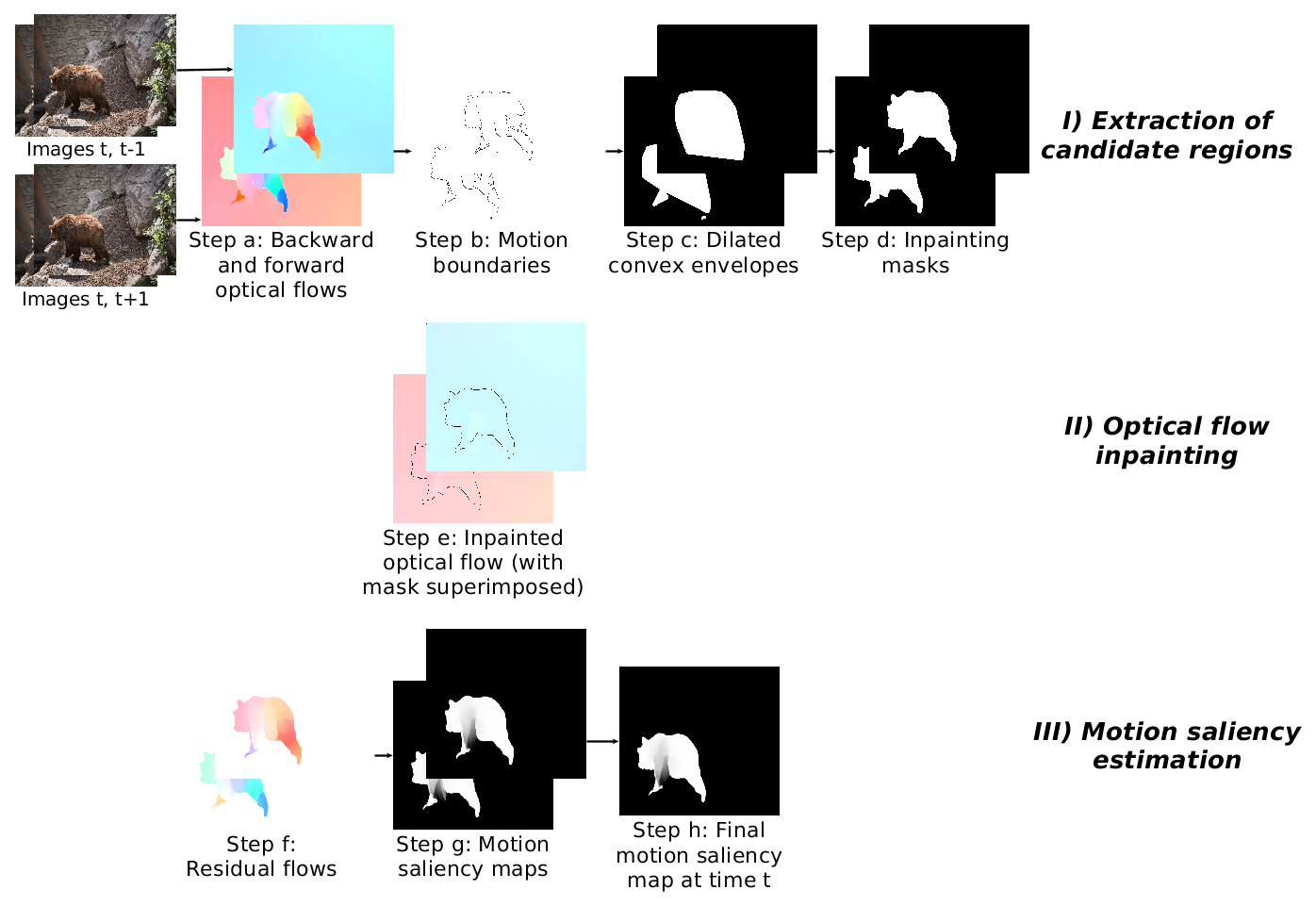 |
Motion saliency map estimation softwareThis software estimates motion saliency maps from video frames. Candidate salient regions are first extracted from motion boundaries. Saliency is then estimated with an optical flow inpainting step. The idea is that if the flow reconstructed in candidate regions from the surrounding flow is very different from the flow present originally, the region should be salient with respect to motion |
 |
Motion saliency detection softwareThis software detects whether motion saliency is present for each frame of a video. In a first step, the dominant motion (that is supposed to correspond to camera motion) is compensated. In a second step, saliency detection is realised with a convolutional neural network. The 4 variants are RFS-Motion2D, RFS-DeepDOM, WS-Motion2D and WS-DeepDOM. |
 |
AiryscanJAiryscanJ is a high resolution image reconstruction from array detector. High resolution images can be reconstructed by using detector linear combinations or deconvolution. Image type: 2D/3D |
 |
AtlasATLAS is a spot detection method. The spots size is automatically selected and the detection threshold adapts to the local image dynamics. ATLAS relies on the Laplacian of Gaussian (LoG) filter, which both reduces noise and enhances spots. A multiscale representation of the image is built to automatically select the optimal LoG variance. Image type: 2D |
 |
BackwarpingWarp sequence with parametric motion model (1995-2013) C/C++ |
 |
C-CRAFTVesicle segmentation and background estimation (2014) C/C++, Java |
 |
Cryo-SegSegmentation of tomograms in cryo-electron microscopy (2010) |
 |
Flowscope3D flow field estimation and assessment for live cell fluorescence microscopy (2019) Image type: 3D + t |
 |
Flux estimationParticle and intensity flux estimation (2015) C/C++ |
 |
GenLoc3D3D high-precision gene localization algorithm in cell nuclei (2014-2015) Java (ImageJ) |
 |
GcoPS3D Colocalization analysis (2017) Java |
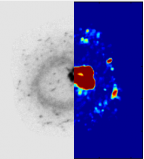 |
HotSpotDetection3D Robust detection of fluorescence accumulation over time in video-microscopy (2007-2008) C/C++ |
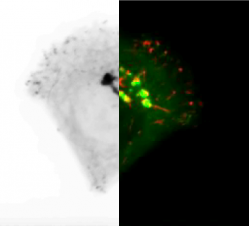 |
HullkGround3D Convex Hull for background subtraction (2008-2009) C/C++, Java |
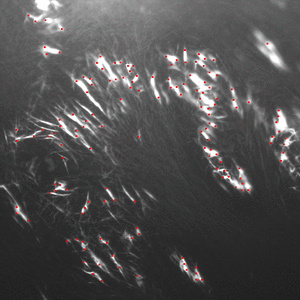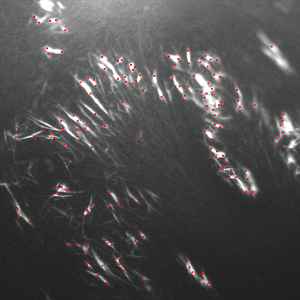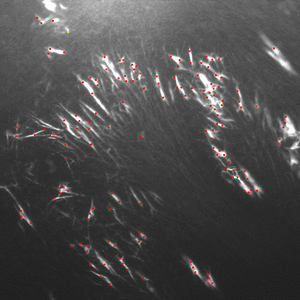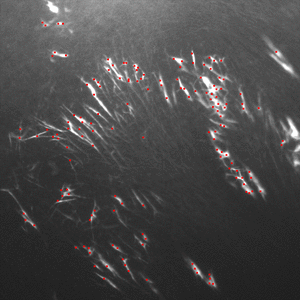 |
KLTrackerTrack vesicle and POI in image sequences (2012) C/C++ |
 |
Motion2DEstimate 2D parametric motion model (2012)C/C++ |
 |
MS-DetectDetecting moving objects in image sequences by background subtraction (2008/2011) C/C++ |
 |
ND-SAFIRND-SAFIR is a software for denoising n-dimentionnal images especially dedicated to microscopy image sequence analysis. It is able to deal with 2D, 3D, 2D+time, 3D+time images have one or more color channel. It is adapted to Gaussian and Poisson-Gaussian noise which are usually encountered in photonic imaging. Several papers describe the detail of the method used in ndsafir to recover noise free images. Image type: 2D, 3D, 2D+time, 3D+time |
 |
OpticalFlowOptical flow (2012) C/C++ |
 |
OWFNon-local means and optimal weights for noise removal (2017) Matlab and C/C++ |
 |
PBEDPatch-Based Event Detection (2009) C/C++ |
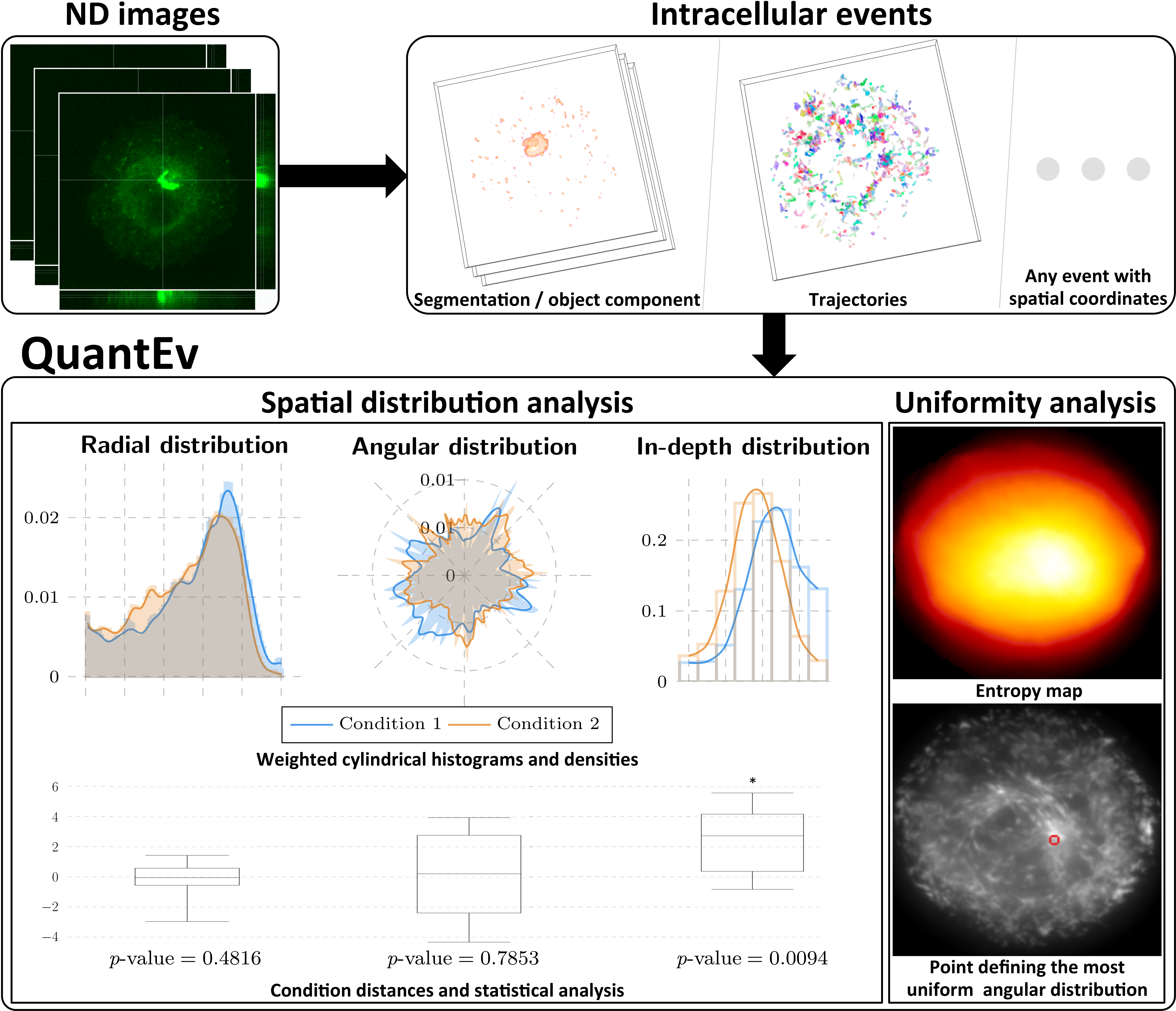 |
QuantEvSpatio-temporal distribution analysis (2017) C/C++, Java |
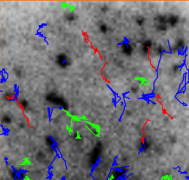 |
THOTHClassification of particle trajectories into three types of diffusions (2017) Matlab |
 |
CPAnalysisChange point detection along particle trajectories (2018) Matlab |
 |
TubuleJToolbox to process electron cryo-microscope images of vitrified microtubules (2009) Java (imageJ) |
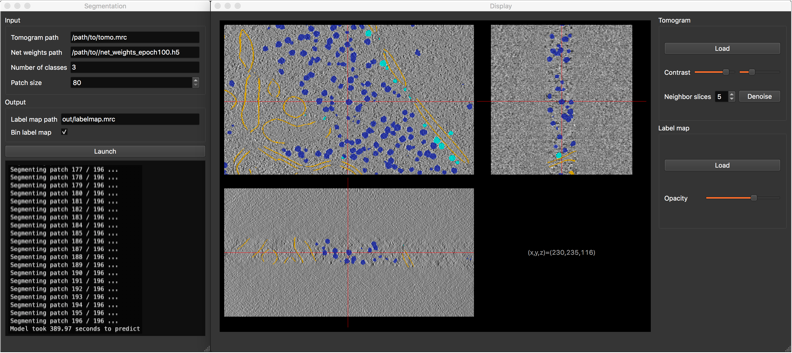 |
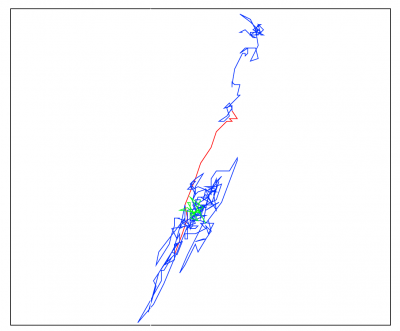
DeepFinderDeepFinder is an original deep learning approach to localize macromolecules in cryo electron tomography images. The method is based on image segmentation using a 3D convolutional neural network. |
 |
DenseMappingDenseMapping software allows to estimate the diffusion and drift parameters attached to moving biomolecules within cells from 2D/3D individual trajectories. |
Demonstrations
Online demonstration of several softwares are available at the Inria Allgo platform: https://allgo.inria.fr/