CPAnalysis software
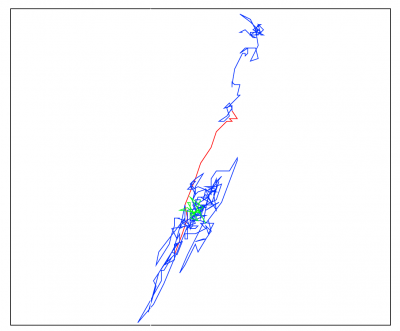
Change point detection along a neuronal mRNP trajectory made with CPAnalysis.
In blue Brownian motion, in green subdiffusion, in red superdiffusion.
Data from Monnier, Nilah, et al. “Inferring transient particle transport dynamics in live cells.” Nature methods 12.9 (2015): 838.
Overview
Our change point detection algorithm is designed for detecting switches of diffusion along single particle trajectories (in both 2D or 3D).
We consider that the particle switches between three main modes of motion :
- Superdiffusion which occurs when the particle is transported via molecular motors along the cytoskeleton.
- Free diffusion (or Brownian motion) which arises when the particle evolves freely inside the cytosol.
- Subdiffusion observed when the particle is confined in a domain or evolves in a crowded area.
The algorithm is a sequential nonparametric procedure based on test statistics computed on local windows along the trajectory.
Software distribution
Matlab source code is freely ditributed under Affero GPL v3.0 license: cpanalysis.zip
Reference
- V. Briane, M. Vimond, C. Valades Cruz, A. Salomon, C. Wunder, C. Kervrann. A sequential algorithm to detect diffusion switching along intracellular particle trajectories, Bioinformatics, btz489, HAL-INRIA-01966831, 2019
- V. Briane, M. Vimond, C. Kervrann. An overview of diffusion models for intracellular dynamics analysis, bbz052, Briefings in Bioinformatics, HAL-INRIA-0196682, 2019