Parietal’s work on modelling brain architecture: works on diffusion MRI, anatomical MRI, and the joint analysis of anatomical and functional features.
NeuroVault.org: a web-based repository for collecting and sharing unthresholded statistical maps of the human brain
NeuroVault is a web based repository that allows researchers to store, share, visualize, and decode statistical maps of the human brain. NeuroVault is easy to use and employs modern web technologies to provide informative visualization of data without the need to install additional software. In addition, it leverages the power of the Neurosynth database to provide cognitive decoding of deposited maps. The data are exposed through a public REST API enabling other services and tools to take advantage of it. NeuroVault is a new resource for researchers interested in conducting meta- and coactivation analyses.

Comparison of image based and coordinate based meta analysis of response inhibition. Meta analysis based on unthresholded statistical maps obtained from NeuroVault (top row) managed to recover the pattern of activation obtained using traditional methods despite including much fewer studies. NeuroVault map has been thresholded at z=6, response inhibition map has been thresholded at z=1.77 (the threshold values were chosen for visualization purposes only, but both are statistically significant at p<0.05). Unthresholded versions of these maps are available at http://neurovault.org/collections/439.
Semi-Supervised Factored Logistic Regression for High-Dimensional Neuroimaging Data
Imaging neuroscience links human behavior to aspects of brain biology in ever-increasing datasets. Existing neuroimaging methods typically perform either discovery of unknown neural structure or testing of neural structure associated with mental tasks. However, testing hypotheses on the neural correlates underlying larger sets of mental tasks necessitates adequate representations for the observations. We therefore propose to blend representation modelling and task classification into a unified statistical learning problem. A multinomial logistic regression is introduced that is constrained by factored coefficients and coupled with an au-toencoder. We show that this approach yields more accurate and interpretable neural models of psychological tasks in a reference dataset, as well as better generalization to other datasets.
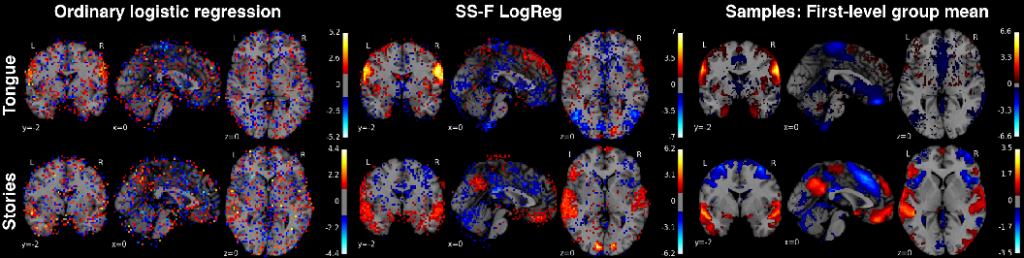
Classification weight maps. The voxel predictors corresponding to 2 exemplary (of 18 total) psychological tasks (rows) from the Human Connectome Project dataset. Left column: multinomial logistic regression (same implementation but without bottleneck or autoencoder), middle column: Semi-Supervised Factored Logistic Regression (SSFLogReg), right column: voxel-wise average across all samples of whole-brain activity maps from each task. SSFLogReg puts higher absolute weights on relevant structure, lowers ones on irrelevant structure, and yields BOLD-typical local con- tiguity (without enforcing an explicit spatial prior).
Principal Component Regression predicts functional responses across individuals
Inter-subject variability is a major hurdle for neuroimaging group-level inference, as it creates complex image patterns that are not captured by standard analysis models and jeopardizes the sensitivity of statistical procedures. A solution to this problem is to model random subjects effects by using the redundant information conveyed by multiple imaging contrasts. In this paper, we introduce a novel analysis framework, where we estimate the amount of variance that is fit by a random effects subspace learned on other images; we show that a principal component regression estimator outperforms other regression models and that it fits a significant proportion (10% to 25%) of the between-subject variability. This proves for the first time that the accumulation of contrasts in each individual can provide the basis for more sensitive neuroimaging group analyzes.
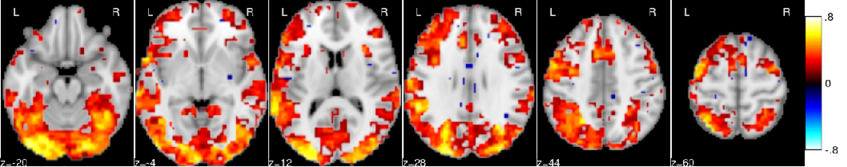
In most brain regions, knowing the amount of activation related to a set of reference contrasts yields an accurate prediction of the activation for a target contrast.
Deriving a multi-subject functional-connectivity atlas to inform connectome estimation
The estimation of functional connectivity structure from functional neuroimaging data is an important step toward understanding the mechanisms of various brain diseases and building relevant biomarkers. Yet, such inferences have to deal with the low signal-to-noise ratio and the paucity of the data. With at our disposal a steadily growing volume of publicly available neuroimaging data, it is however possible to improve the estimation procedures involved in connectome mapping. In this work, we propose a novel learning scheme for functional connectivity based on sparse Gaussian graphical models that aims at minimizing the bias induced by the regularization used in the estimation, by carefully separating the estimation of the model support from the coefficients. Moreover, our strategy makes it possible to include new data with a limited computational cost. We illustrate the physiological relevance of the learned prior, that can be identified as a functional connectivity atlas, based on an experiment on 46 subjects of the Human Connectome Dataset.
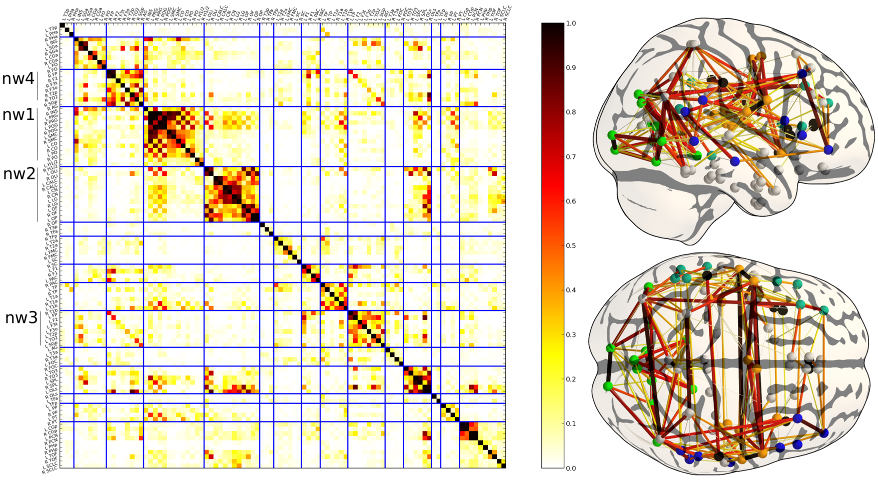
Prior on the functional connectivity: the coefficient of the matrix represent the frequency of an edge at each position. This model can be interpreted as a data-driven atlas of brain functional connections. In the current framework, it can easily be updated to take into account more data.
Which fMRI clustering gives good brain parcellations?
Analysis and interpretation of neuroimaging data often require one to divide the brain into a number of regions, or parcels, with homogeneous characteristics, be these regions defined in the brain volume or on on the cortical surface. While predefined brain atlases do not adapt to the signal in the individual subjects images, parcellation approaches use brain activity (e.g. found in some functional contrasts of interest) and clustering techniques to define regions with some degree of signal homogeneity. In this work, we address the question of which clustering technique is appropriate and how to optimize the corresponding model. We use two principled criteria: goodness of fit (accuracy), and reproducibility of the parcellation across bootstrap samples. We study these criteria on both simulated and two task-based functional Magnetic Resonance Imaging datasets for the Ward, spectral and K-means clustering algorithms. We show that in general Ward’s clustering performs better than alternative methods with regard to reproducibility and accuracy and that the two criteria diverge regarding the preferred models (reproducibility leading to more conservative solutions), thus deferring the practical decision to a higher level alternative, namely the choice of a trade-off between accuracy and stability.
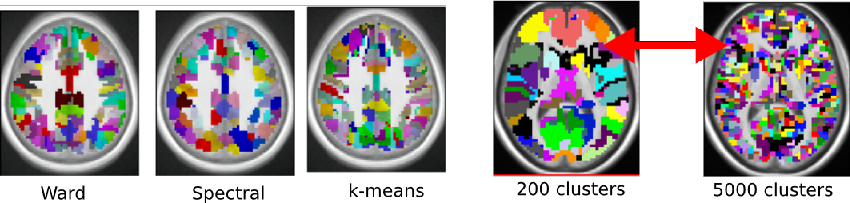
Practitioner have to decide which clustering method to use and how to select the number of clusters. We provide empirical guidelines and criteria to guide that choice in the context of functional brain imaging.
Mapping cognitive ontologies to and from the brain
Imaging neuroscience links brain activation maps to behavior and cognition via correlational studies. Due to the nature of the individual experiments, based on eliciting neural response from a small number of stimuli, this link is incomplete, and unidirectional from the causal point of view. To come to conclusions on the function implied by the activation of brain regions, it is necessary to combine a wide exploration of the various brain functions and some inversion of the statistical inference. Here we introduce a methodology for accumulating knowledge towards a bidirectional link between observed brain activity and the corresponding function. We rely on a large corpus of imaging studies and a predictive engine. Technically, the challenges are to find commonality between the studies without denaturing the richness of the corpus. The key elements that we contribute are labeling the tasks performed with a cognitive ontology, and modeling the long tail of rare paradigms in the corpus. To our knowledge, our approach is the first demonstration of predicting the cognitive content of completely new brain images. To that end, we propose a method that predicts the experimental paradigms across different studies (see Fig. below ).
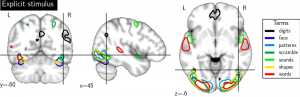
Maps for the forward inference for some cognitive category related to the semantic content of brain images.
Cohort-level brain mapping: learning cognitive atoms to single out specialized regions
Functional Magnetic Resonance Imaging (fMRI) studies map the human brain by testing the response of groups of individuals to carefully-crafted and contrasted tasks in order to delineate specialized brain regions and networks. The number of functional networks extracted is limited by the number of subject-level contrasts and does not grow with the cohort. Here, we introduce a new group-level brain mapping strategy to differentiate many regions reflecting the variety of brain network configurations observed in the population. Based on the principle of functional segregation, our approach singles out functionally-specialized brain regions by learning group-level functional profiles on which the response of brain regions can be represented sparsely. We use a dictionary-learning formulation that can be solved efficiently with on-line algorithms, scaling to arbitrary large datasets. Importantly, we model inter-subject correspondence as structure imposed in the estimated functional profiles, integrating a structure-inducing regularization with no additional computational cost. On a large multi-subject study, our approach extracts a large number of brain networks with meaningful functional profiles (see Fig. below).
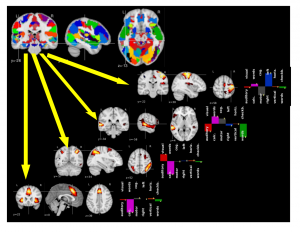
A brain functional atlas can be conceptualized as a parcellation of the brain volume into overlapping networks, where each functional network is characterized by a profile of activation for a set of functional contrasts.
Multi-subject volume- vs surface-based fMRI data analysis
Being able to detect reliably functional activity in a population of subjects is crucial in human brain mapping, both for the understanding of cognitive functions in normal subjects and for the analysis of patient data.
The usual approach proceeds by normalizing brain volumes to a common 3D template. However, a large part of the data acquired in fMRI aims at localizing cortical activity, and methods working on the cortical surface may provide better inter-subject registration than the standard procedures that process the data in 3D. Nevertheless, few assessments of the performance of surface-based (2D) versus volume-based (3D) procedures have been shown so far, mostly because inter-subject cortical surface maps are not easily
obtained.
We have presented in Tucholka:09 a systematic comparison of 2D versus 3D group-level inference procedures, by using cluster-level and voxel-level statistics assessed by permutation, in random effects (RFX) and mixed-effects analyses (MFX). We found that, using a voxel-level thresholding, and to some extent, cluster-level thresholding, the surface-based approach generally
detects more, but smaller active regions than the corresponding volume-based approach for both RFX and MFX procedures (see Fig. below). Finally we showed that surface-based supra-threshold regions are more
reproducible by bootstrap.


