Accessing cytoarchitecture in human brain gray matter via a forward diffusion signal separation method
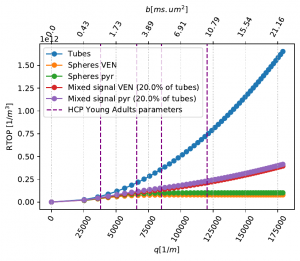
Our contribution focuses on the human brain gray matter, which can be decomposed into three compartments (somas, processes, and cerebrospinal fluid) and modeled by simple shapes : spheres and 0-radius tubes. We characterize and quantify the diffusion MRI signal coming from those micrometer-thin cells. Studying the restrictions of the diffusing fluid molecule motion gives us direct information about the structure of the media. This can be achieved by computing the return-to-the-origin probability, which is the probability that diffusing molecules will return to (or have not left) the vicinity of their initial positions after a given diffusion time. Hence it provides a quantity related to the structure of the media, i.e. local properties for short diffusion times and the connections and restrictions of the complex neural structure for long diffusion times. We then suggest a forward model to separate the contributions of somas and processes and avoid degeneracy implied by inverse models.
Tests are run on simulations using the DMIPY library and on the HCP1200 data set, a freely and high-quality neuroimaging data.

 [Palombo et al., 2017]
[Palombo et al., 2017]
References
Jallais, M. & Wassermann, D.
Indetermination-free cytoarchitecture measurements in brain gray matter via a forward diffusion MRI signal separation method
International Society for Magnetic Resonance in Medicine (ISMRM) (2020)
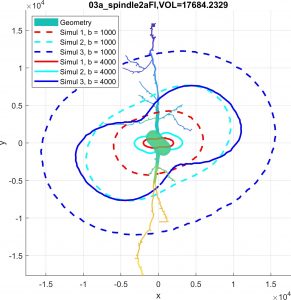
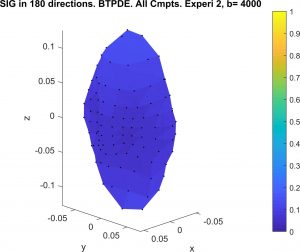
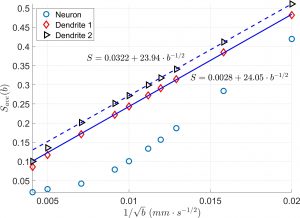
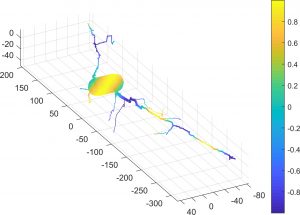
Diffusion MRI simulation of realistic neurons with SpinDoctor and the Neuron Module
In collaboration with DEFI team (Inria).
Diffusion MRI is an imaging modality that plays an important role in the tractography of the human brain’s white matter. A preliminary work of our team has shown that the presence of spindle neurons induces an attenuation to the dMRI signal even in clinical settings. This provides a promising way of dMRI-based in-vivo microscopy of the human brain. Some microstructure parameters such as axon diameter, orientation, neurite density can also be detected by dMRI with well-designed acquisition protocols and processing algorithms. A dMRI simulator, called SpinDoctor, recently proposed by our lab is proved to be 100 times faster than a Monte-Carlo based simulator of Camino. This huge improvement of simulation efficiency allows us to further investigate the relation between the brain microstructure and the dMRI signal.