Work at Parietal on statistical analysis for multi subject analysis of neuroimaging data, and the comparison of neuroimaging data with genetic or behavioural information.
Multi-subject MEG/EEG source localization with Optimal transport
EEG/MEG source localization is an ill-conditioned linear inverse problem. One way to improve its conditionning is to solve it for multiple subjects with a binding regularization. This corresponds to a multi-task regression problem where the regularizer induces some similarity across the regression coefficients. In the context of source localization, adding a similarity prior means promoting functional consistency across subjects. This assumption
can be encountered when studying evoked responses of the same cognitive task. Moreover, to cope with the underdetermined nature of the inverse problems, one can favor sparse active regions over distributed ones using sparse regularizers. We studied several multi-task models using the Cam-CAN dataset; our main conclusions are twofold: different brain anatomies (different regression matrices or leadfields X) lead to better source recovery; Optimal transport models capture spatial variability and can thus promote functional consistency in a flexible manner. Details of our study can be found in this paper .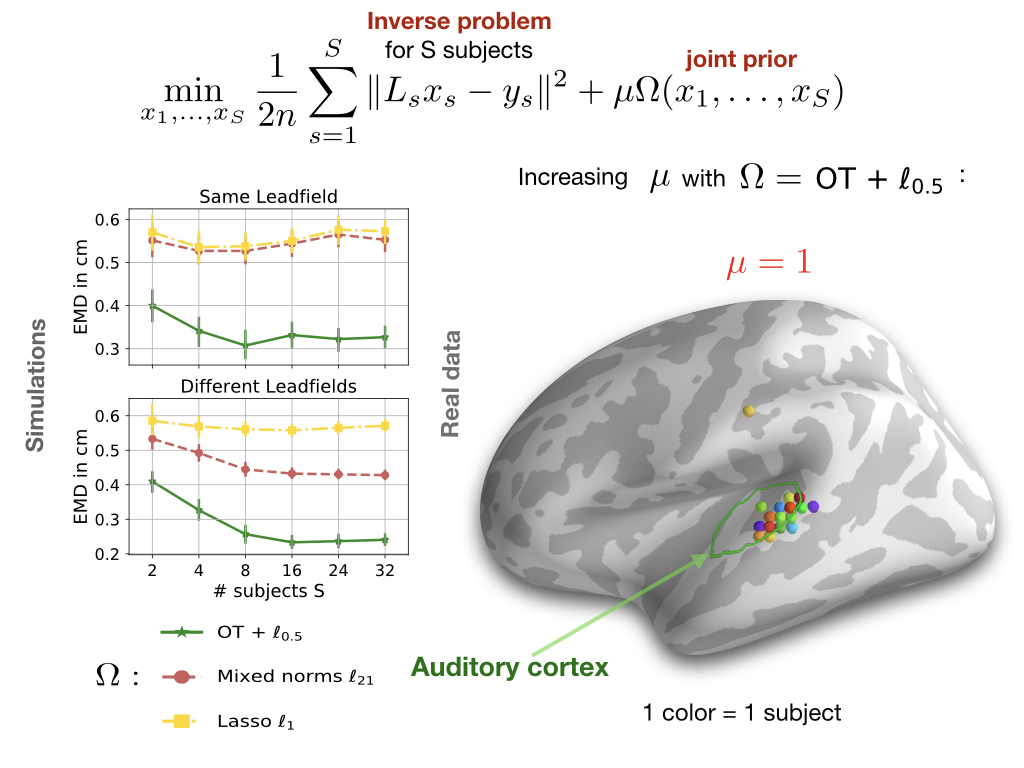
Integrating Multimodal Priors in Predictive Models for the Functional Characterization of Alzheimer’s Disease
Functional brain imaging provides key information to characterize neurodegenerative diseases, such as Alzheimer’s disease (AD). Specifically, the metabolic activity measured through fluorodeoxyglucose positron emission tomography (FDG-PET) and the connectivity extracted from resting-state functional magnetic resonance imaging (fMRI), are promising biomarkers that can be used for early assessment and prognosis of the disease and to understand its mechanisms. FDG-PET is the best suited functional marker so far, as it gives a reliable quantitative measure, but is invasive. On the other hand, non-invasive fMRI acquisitions do not provide a straightforward quantification of brain
functional activity. To analyze populations solely based on resting-state fMRI, we propose an approach that leverages a metabolic prior learned from FDG-PET. More formally, our classification framework embeds population priors learned from another modality at the voxel-level, which can be seen as a regularization
term in the analysis. Experimental results show that our PET-informed approach increases classification accuracy compared to pure fMRI approaches and highlights regions known to be impacted by the disease.

Overview of the proposed classification pipeline: The inputs are ROI-to-voxel connectivities computed from the rs-fMRI time-series. FDG-PET model weights are integrated as prior for the classification. Then, predictions of all ROIs are the inputs of a stacking model to predict the clinical group.
Improving sparse recovery on structured images with bagged clustering
The identification of image regions associated with external variables through discriminative approaches yields ill-posed estimation problems. This estimation challenge can be tackled by imposing sparse solutions. However, the sensitivity of sparse estimators to correlated variables leads to non-reproducible results, and only a subset of the important variables are selected. In this paper, we explore an approach based on bagging clustering-based data compression in order to alleviate the instability of sparse models. Specifically, we design a new framework in which the estimator is built by averaging multiple models estimated after feature clustering, to improve the conditioning of the model. We show that this combination of model averaging with spatially consistent compression can have the virtuous effect of increasing the stability of the weight maps, allowing a better interpretation of the results. Finally, we demonstrate the benefit of our approach on several predictive modeling problems.
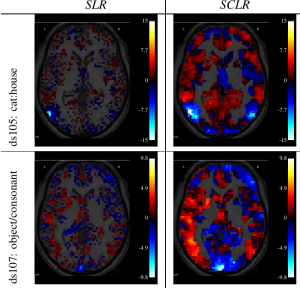
Z-score obtained across bootstraps for two discriminative tasks, using Sparse logistic Regression without (SLR) and with (SCLR) clustering. Higher values hint at lower variability across bootstrap replications. SCLR decreases the variability and yields
larger standardized effects.
Bootstrapped Permutation Test for Multiresponse Inference on Brain Behavior Associations.
Despite that diagnosis of neurological disorders commonly involves a collection of behavioral assessments, most neuroimaging studies investigating the associations between brain and behavior largely analyze each behavioral measure in isolation. To jointly model multiple behavioral scores, sparse mul-tiresponse regression (SMR) is often used. However, directly applying SMR without statistically controlling for false positives could result in many spurious findings. For models, such as SMR, where the distribution of the model parameters is unknown, permutation test and stability selection are typically used to control for false positives. In this paper, we present another technique for inferring statistically significant features from models with unknown parameter distribution. We refer to this technique as bootstrapped permutation test (BPT), which uses Studentized statistics to exploit the intuition that the variability in parameter estimates associated with relevant features would likely be higher with responses permuted. On synthetic data, we show that BPT provides higher sensitivity in identifying relevant features from the SMR model than permutation test and stability selection, while retaining strong control on the false positive rate. We further apply BPT to study the associations between brain connec-tivity estimated from pseudo-rest fMRI data of 1139 fourteen year olds and be-havioral measures related to ADHD. Significant connections are found between brain networks known to be implicated in the behavioral tasks involved. Moreover , we validate the identified connections by fitting a regression model on pseudo-rest data with only those connections and applying this model on resting state fMRI data of 337 left out subjects to predict their behavioral scores. The predicted scores are shown to significantly correlate with the actual scores of the subjects, hence verifying the behavioral relevance of the found connections.
 Real data results: Statistically significant connectivity differences between populations (a) Significant network connections found on pseudo-rest fMRI data. (b) Pearson’s correlation between predicted and actual scores with p-values noted. Each set of three bars (top to bottom) correspond to spatial working memory strategy, spatial working memory between errors, and rapid visual information processing accuracy scores. Significance is declared at p <.05.
Real data results: Statistically significant connectivity differences between populations (a) Significant network connections found on pseudo-rest fMRI data. (b) Pearson’s correlation between predicted and actual scores with p-values noted. Each set of three bars (top to bottom) correspond to spatial working memory strategy, spatial working memory between errors, and rapid visual information processing accuracy scores. Significance is declared at p <.05.
Machine Learning Patterns for Neuroimaging-Genetic Studies in the Cloud
Brain imaging is a natural intermediate phenotype to understand the link between genetic information and behavior or brain pathologies risk factors. Massive efforts have been made in the last few years to acquire high-dimensional neuroimaging and genetic data on large cohorts of subjects. The statistical analysis of such data is carried out with increasingly sophisticated techniques and represents a great computational challenge. Fortunately, increasing computational power in distributed architectures can be harnessed, if new neuroinformatics infrastructures are designed and training to use these new tools is provided. Combining a MapReduce framework (TomusBLOB) with machine learning algorithms (Scikit-learn library), we design a scalable analysis tool that can deal with non-parametric statistics on high-dimensional data. End-users describe the statistical procedure to perform and can then test the model on their own computers before running the very same code in the cloud at a larger scale. We illustrate the potential of our approach on real data with an experiment showing how the functional signal in subcortical brain regions can be significantly fit with genome-wide genotypes. This experiment demonstrates the scalability and the reliability of our framework in the cloud with a two weeks deployment on hundreds of virtual machines.
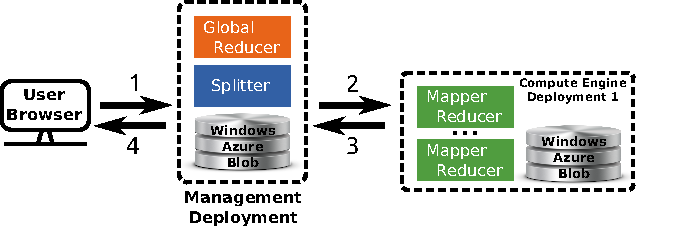
Overview of the multi site deployment of a hierarchical Tomus-MapReduce compute engine. 1) The end-user uploads the data and configures the statistical inference procedure on a webpage. 2) The Splitter partitions the data and manages the workload. The compute engines retrieves job information trough the Windows Azure Queues. 3) Compute engines perform the map and reduce jobs. The management deployment is informed of the progression via the Windows Azure Queues system and thus can manage the execution of the global reducer. 4) The user downloads the results of the computation on the webpage of the experiment.
Group-level impacts of within- and between-subject hemodynamic variability in fMRI
Inter-subject fMRI analyses have specific issues regarding the reliability of the results concerning both the detection of brain activation patterns and the estimation of the underlying dynamics. Among these issues lies the variability of the hemodynamic response function (HRF), that is usually accounted for using functional basis sets in the general linear model context. Here, we use the joint detection-estimation approach (JDE) [76] , [78] , which combines regional nonparametric HRF inference with spatially adaptive regularization of activation clusters to avoid global smoothing of fMRI images (see Fig. 4 ). We show that the JDE-based inference brings a significant improvement in statistical sensitivity for detecting evoked activity in parietal regions. In contrast, the canonical HRF associated with spatially adaptive regularization is more sensitive in other regions, such as motor cortex. This different regional behavior is shown to reflect a larger discrepancy of HRF with the canonical model. By varying parallel imaging acceleration factor, SNR-specific region-based hemodynamic parameters (activation delay and duration) were extracted from the JDE inference. Complementary analyses highlighted their significant departure from the canonical parameters and the strongest between-subject variability that occurs in the parietal region, irrespective of the SNR value. Finally, statistical evidence that the fluctuation of the HRF shape is responsible for the significant change in activation detection performance is demonstrated using paired t-tests between hemodynamic parameters inferred by GLM and JDE.
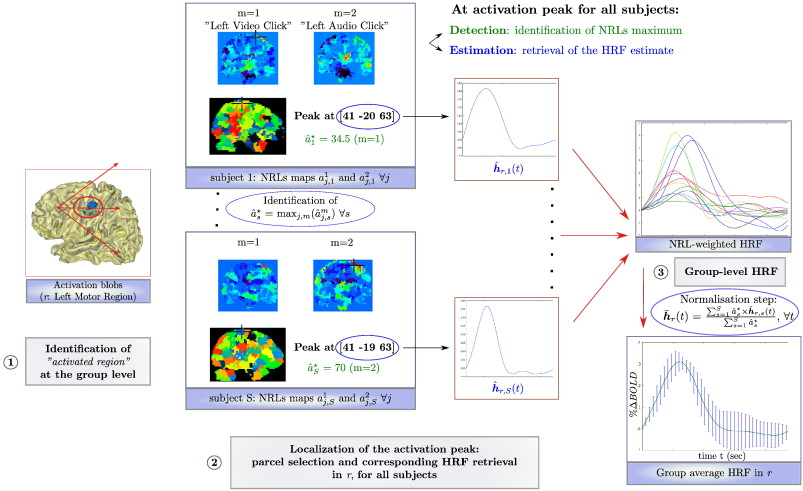
General sketch summarizing the HRF computation at the subject and group-levels in activated regions r. Left: Position of the activation peak in r (here left motor cortex) given in mm in the Talairach space. Center: Individual weighted HRF time course extraction. Right: Computation of the group average normalized HRF time course with corresponding error bars (±σ).
Randomized parcellation-based inference
Neuroimaging group analyses are used to relate inter-subject signal differences observed in brain imaging with behavioral or genetic variables and to assess risks factors of brain diseases. The lack of stability and of sensitivity of current voxel-based analysis schemes may however lead to non-reproducible results. We introduce a new approach to overcome the limitations of standard methods, in which active voxels are detected according to a consensus on several random parcellations of the brain images, while a permutation test controls the false positive risk (see Fig. below). Both on synthetic and real data, this approach shows higher sensitivity, better accuracy and higher reproducibility than state-of-the-art methods. In a neuroimaging-genetic application, we find that it succeeds in detecting a significant association between a genetic variant next to the COMT gene and the BOLD signal in the left thalamus for a functional Magnetic Resonance Imaging contrast associated with incorrect responses of the subjects from a Stop Signal Task protocol.
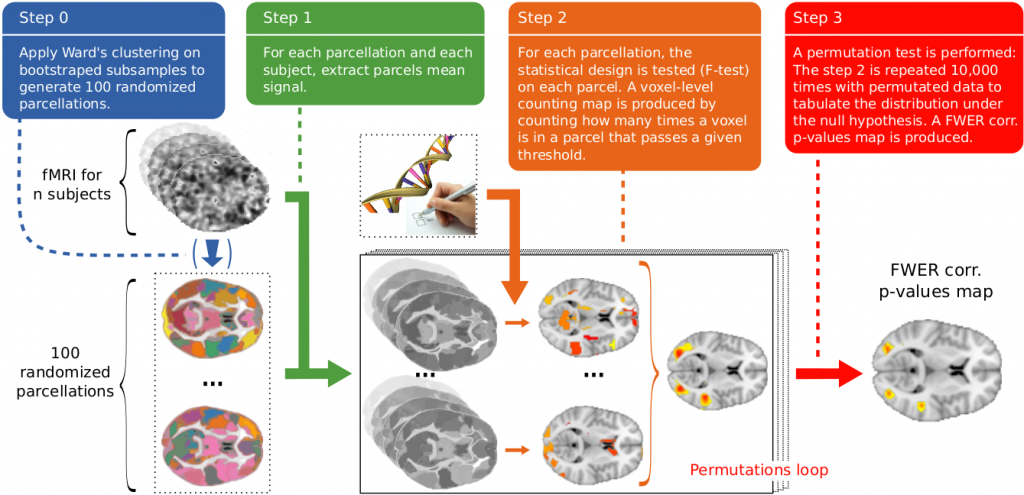
Overview of the randomized parcellation based inference framework on an example with few parcels. The variability of the parcels definition is used to obtain voxel-level statistics.
Parcel-based random effects analyses
Presented at COMPSTAT 2010
Activation detection in functional Magnetic Resonance Imaging (fMRI) datasets is usually performed by thresholding activation maps in the brain volume or, better, on the cortical surface. However, basing the analysis on a site-by-site statistical decision may be detrimental both to the interpretation of the results and to the sensitivity of the analysis, because a perfect point-to-point correspondence of brain surfaces from multiple subjects cannot be guaranteed in practice. In this work, we propose a new approach that first defines anatomical regions such as cortical gyri outlined on the cortical surface, and then segments these regions into functionally homogeneous structures using a parcellation procedure that includes an explicit between-subject variability model, i.e. random effects. We show that random effects inference can be performed in this framework. Our procedure allows an exact control of the specificity using permutation techniques, and we show that the sensitivity of this approach is higher than the sensitivity of voxel- or cluster-level random effects tests performed on the cortical surface.

Outcome of the cluster-based (left), parcel-based (middle) and node-based (right) random effects analyses in the left(top) and right (bottom) hemisphere. All the maps are corrected at the p
ICOGEN : Intensive COmputing for GEnetic-Neuroimaging studies
More details on the dedicated page
In this project, we design and deploy some computational tools to perform neuroimaging-genetics association studies at a large scale.
HiDiNim: High-dimensional Neuroimaging — Statistical Models of Brain Variability observed in Neuroimaging
More details on the dedicated page
In this work, we propose to investigate the statistical structure of large populations observed in neuroimaging. In particular, we will investigate the use of region-level averages of brain activity, that we plan to co-analyse with genetic and behavioral information, in order to understand the sources of the observed variability.