MICROCOSME combines computational and experimental approaches for the analysis, engineering, and control of the growth of micro-organisms. The team joins researchers from Inria Grenoble – Rhône-Alpes and the Laboratoire Interdisciplinaire de Physique at Université Grenoble Alpes (CNRS UMR 5588).
Microbial growth
Microorganisms are found everywhere, in soils, in water, and in humans, where on average 1.3 bacterial cells are counted for every human cell. One of the defining characteristics of microorganisms is their capacity to grow and divide, that is, transform nutrients from the environment into new microbial cells. They multiply fast in permissive conditions (doubling every twenty minutes), slow in adverse conditions (doubling every few hours or days), or not at all in case of extreme stress. Understanding and controlling the dynamics of bacterial growth is vitally important in health, medicine, biotechnology, and food industries, for instance to halt the growth of pathogens or stimulate the growth of probiotics or industrial microorganisms.
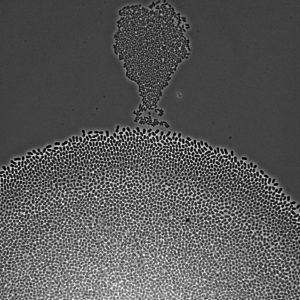
Microscopy image of Escherichia coli bacteria growing on a solid nutrient medium. Some bacteria have grown in the form of a hot air balloon (top) which, by colonizing the surface, will soon fuse with a second, bigger colony (bottom). The bacteria are rod shaped, 2 µm long, and divide every 20 minutes in the conditions in which the picture was taken.
Credit: Antrea Pavlou, Dec 2020.
The inside story
Microbial growth is essentially a multiscale phenomenon, in the sense that the macroscopic observable, growth of a microbial population, depends on various metabolic pathways and regulatory mechanisms operating at microscopic scales within the cells. Novel experimental technologies have provided an increasingly detailed and quantitative account of these pathways and regulatory mechanisms, including complex feedback mechanisms operating on different time-scales. Recent advances in genome engineering have used this knowledge to enable directed genetic modifications that turn microorganisms into microbial cell factories capable of producing a range of metabolites, peptides, and proteins of industrial and medical interest.
Despite these technological advances, many if not most fundamental questions on microbial growth remain open. For instance, growth of microorganisms requires the synthesis of different cellular components in a timely manner under resource constraints. How do cells coordinate the different intracellular growth processes and adjust bacterial growth rate to an ever-changing environment? How do they allocate limiting cellular machineries to these processes so as to grow optimally in permissive conditions or to survive in harmful environments? How do fluctuations lead to heterogeneous growth behaviour over a population? How can feedback control stabilize the composition of a synthetic community of microorganisms and optimize the production of a metabolite of interest?
Research in MICROCOSME
Research in MICROCOSME addresses these questions. The research program is organized around four axes depicted below, which make use of tools from mathematical modelling (stochastic and deterministic), dynamical systems analysis, control theory, genome and bioprocess engineering:
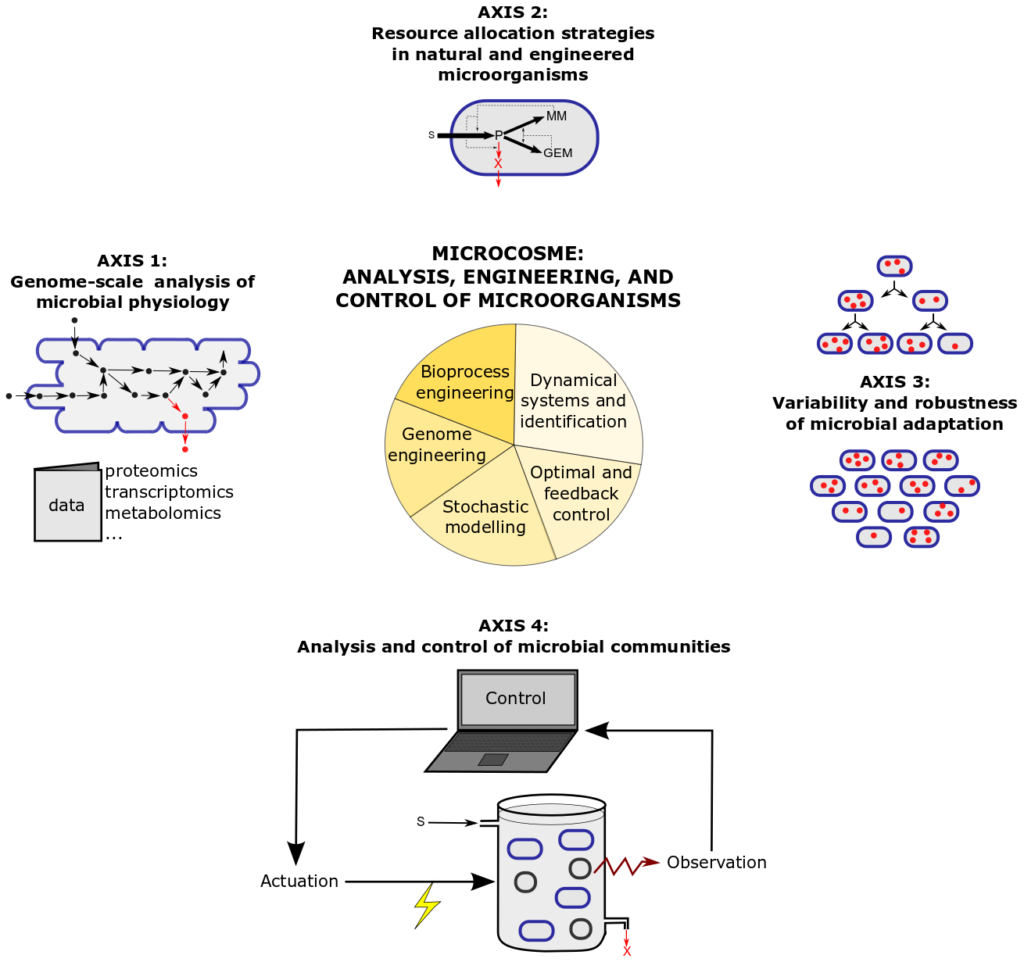
Research axes and methods in MICROCOSME. The first axis is dedicated to a genome-scale understanding of microbial physiology through model-based analysis of high-throughput data. This
allows us to comprehend how cells adjust growth processes to environmental perturbations. This coordination reflects strategies evolved by microorganisms to allocate their resources over different cellular functions to (optimally) grow in their environment. The study of these natural strategies and their re-engineering is the focus of Research axis 2 which views cells as self-replicators that can be described using coarse-grained models and analysed by means of optimal and feedback control theory. In Research axis 3, we adopt a different angle by analysing the variability and robustness of microbial growth. In particular, we shift from deterministic to stochastic models, using data on the level of single cells in a population rather than averaged over all cells in the population. In Research axis 4, a different type of variability is considered, namely heterogeneity within communities consisting of different microbial species and how the community can be controlled for biotechnological applications. Research carried out in the four research axes relies on the methodological resources described in the pie at the centre of the figure.

