Genetic Network Analyzer (GNA) is a computer tool for the modeling and simulation of genetic regulatory networks. The aim of GNA is to assist biologists in constructing a model of a genetic regulatory network using knowledge about regulatory interactions in combination with gene expression data.
Genetic Network Analyzer consists of a simulator of qualitative models of genetic regulatory networks in the form of piecewise-linear differential equations. Instead of exact numerical values for the parameters, which are often not available for networks of biological interest, the user of GNA specifies inequality constraints. This information is sufficient to generate a state transition graph that describes the qualitative dynamics of the network. The simulator has been implemented in Java and has been applied to the analysis of various regulatory systems, such as the networks controlling the initiation of sporulation in B. subtilis, the carbon starvation response in E. coli, and biodegradation of polluants by P. putida. See the list of publications below for references to these and other examples.
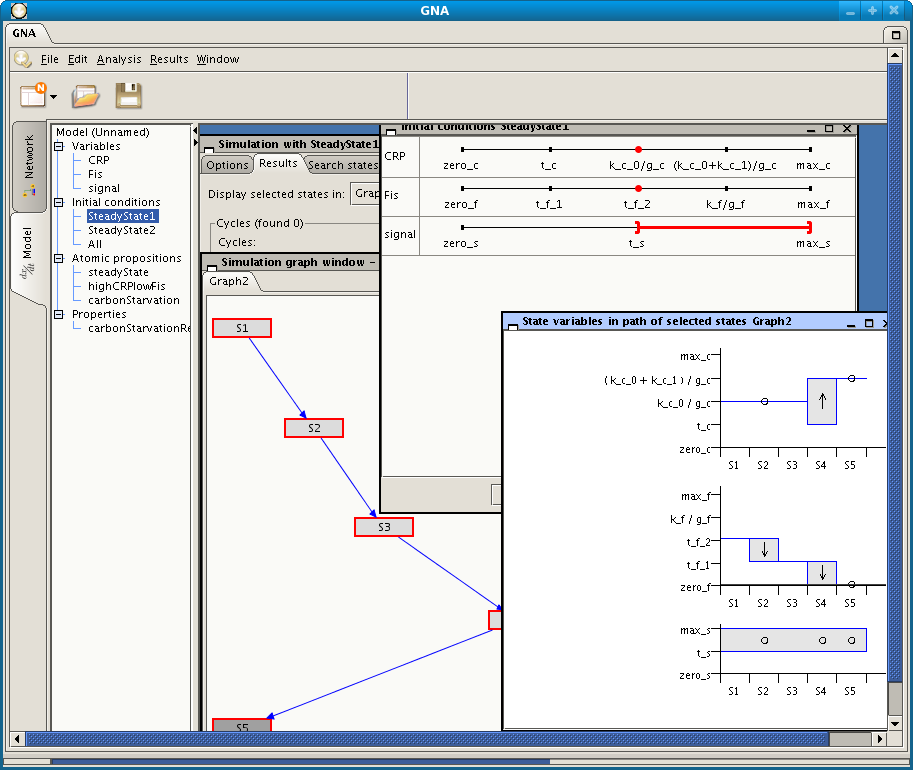
Screenshot of GNA
Functionalities of GNA
The current version is GNA 8.7. In comparison with the previously distributed versions, GNA 8.7 has the following additional functionalities :
- The export of models to the SBML Qual format.
GNA 8.7 preserves all functionalities of previous versions, including :
- The construction of a model using a graphical user interface;
- The performance of a qualitative simulation of the network dynamics, resulting in predictions adapted to available gene expression data;
- The location of all steady states of the model, including an indication of their stability;
- The export and import of models in SBML format;
- The specification of the qualitative dynamics of a network in temporal logic, using high-level query templates;
- The automated verification of these properties on the state transition graph by means of standard model-checking tools, either locally installed or accessible through a remote web server;
- The editing and visualization of regulatory networks, in an SBGN-compatible format;
- The semi-automatic generation of a prototype model from the network structure.
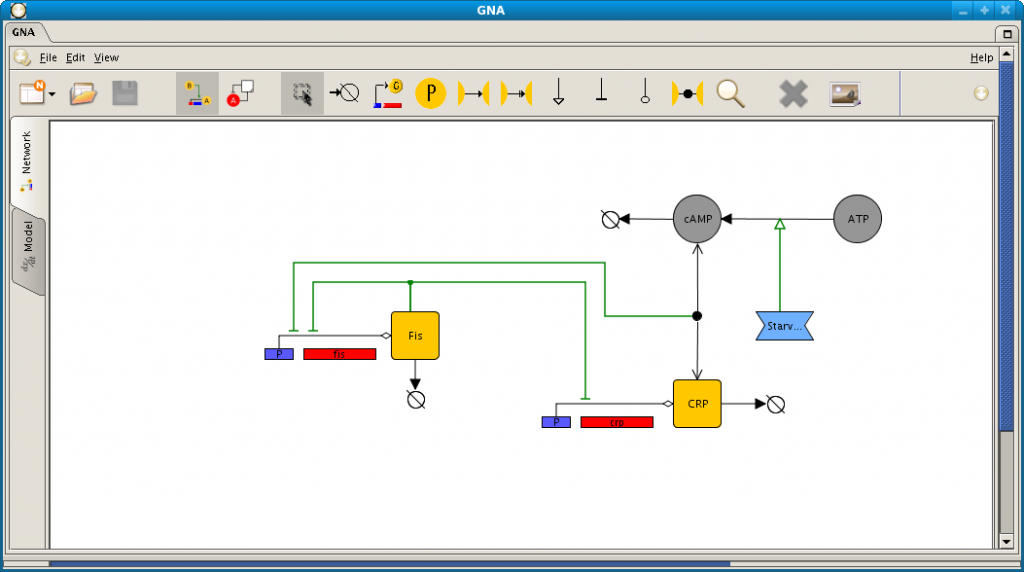
Screenshot of network and model editor in GNA
Obtaining and using Genetic Network Analyzer
Genetic Network Analyzer has been developed in collaboration with the company Genostar, which has integrated GNA in its environment for exploratory genomics, called IOGMA. GNA is freely available for non-profit academic research and the executables can be downloaded for installation here:
Non-academic users wishing to use GNA – or academic users wishing to use GNA for purposes not covered by the user licence – are invited to contact Hidde de Jong.
More information on how to use GNA can be found in the tutorial and the installation guide.
Developers of Genetic Network Analyzer
The main developers of GNA are Hidde de Jong and Michel Page, with important contributions from Grégory Batt, Bruno Besson, Estelle Dumas, Céline Hernandez, Radu Mateescu, and Pedro Monteiro in the past.
References
- H. de Jong, J. Geiselmann, C. Hernandez, M. Page. Genetic Network Analyzer: Qualitative simulation of genetic regulatory networks. Bioinformatics, 19(3):336-344, 2003
- H. de Jong, J. Geiselmann, G. Batt, C. Hernandez, M. Page. Qualitative simulation of the initiation of sporulation in Bacillus subtilis. Bulletin of Mathematical Biology, 66(2):261-300, 2004
- A. Viretta, M. Fussenegger. Quorum sensing in Pseudomonas aeruginosa. Biotechnology Progress, 20(3):670-8, 2004
- G. Batt, D. Ropers, H. de Jong, J. Geiselmann, R. Mateescu, M. Page, D. Schneider. Validation of qualitative models of genetic regulatory networks by model checking: analysis of the nutritional stress response in Escherichia coli. Bioinformatics, 21(Suppl 1):i19-28, 2005. Special issue ISMB 2005
- D. Ropers, H. de Jong, M. Page, D. Schneider, J. Geiselmann. Qualitative simulation of the carbon starvation response in Escherichia coli. Biosystems, 84 (2):124-152, 2006
- J. Sepulchre, S. Reverchon, W. Nasser. Onset of virulence in Erwinia chrysanthemi. Journal of Theoretical Biology, 244(2):239-57, 2007
- H. de Jong, M. Page. Search for steady states of piecewise-linear differential equation models of genetic regulatory networks. ACM/IEEE Transactions on Computational Biology and Bioinformatics, 5(2):508-522, 2008
- P.T. Monteiro, D. Ropers, R. Mateescu, A.T. Freitas, H. de Jong. Temporal logic patterns for querying dynamic models of cellular interaction networks. Bioinformatics, 24(16) :i227-i233. Special issue ECCB 2008
- P.T. Monteiro, E. Dumas, B. Besson, R. Mateescu, M. Page, A.T. Freitas, H. de Jong. A service-oriented architecture for integrating the modeling and formal verification of genetic regulatory networks. BMC Bioinformatics, 10:450, 2009
- G. Batt, M. Page, I. Cantone, G. Goessler, P. Monteiro, H. de Jong. Efficient parameter search for qualitative models of regulatory networks using symbolic model checking. Bioinformatics, 26(18):i603-i610, 2010. Special issue ECCB 2010
- R. Silva-Rocha, H. de Jong, J. Tamames, V. de Lorenzo. The logic layout of the TOL network of Pseudomonas putida pWW0 plasmid stems from a metabolic amplifier motif (MAM) that optimizes biodegradation of m-xylene. BMC Systems Biology, 5:191, 2011
- P.T. Monteiro, P.J. Dias, D. Ropers, A.L. Oliveira, I. Sá-Correia, M.C. Teixeira, A.T. Freitas. Qualitative modeling and formal verification of the FLR1 gene mancozeb response in Saccharomyces cerevisiae. IET Systems Biology, 5(5):308-316, 2011
- G. Batt, B. Besson, P.-E. Ciron, H. de Jong, E. Dumas, J. Geiselmann, R. Monte, P.T. Monteiro, M. Page, F. Rechenmann, D. Ropers. Genetic Network Analyzer: A tool for the qualitative modeling and simulation of bacterial regulatory networks. In: J. van Helden, A. Toussaint, D. Thieffry (eds), Bacterial Molecular Networks, Methods in Molecular Biology, Humana Press, Springer, New York, 2012, 439-462
- A. Métris, S.M. George, D. Ropers. Piecewise linear approximations to model the dynamics of adaptation to osmotic stress by food-borne pathogens. International Journal of Food Microbiology, 240:63-74, 2017
- M. Chaves, H. de Jong. Qualitative modeling, analysis and control of synthetic regulatory circuits. In: F. Menolascine (ed.), Synthetic Gene Circuits: Methods and Protocols, Methods in Molecular Biology 2229, Humana Press, New York, 2021, 1-40
Acknowledgments
GNA uses the following open-source software : JGraph, SAT4J, and CUP. The model-checker web server at INRIA runs NuSMV.

