QUANTIC – QUANTitative modeling combined to statistical learning to understand and predict resistance to Immune-checkpoint inhibition in non-small cell lung Cancer
P.I. : S. Benzekry
COMPO members involved : L Greillier, J. Ciccolini, P. Dufossé, C. Bigarre, S. Marolleau, M. Boussena
Partners : Clinicians: P. Tomasini (AP-HM), F. Barlesi (IGR), Institutional: AP-HM, Inserm, CNRS, Marseille Immunopole, CIML. Researchers: D. Olive (CRCM), E. Vivier/F. Vély (CIML), F. Sabatier (AP-HM), Industrials: Halio DX/Veracyte, Innate Pharma.
Clinical need
The rise of Immune Checkpoint Inhibitors (ICI) has provided significant improvement in survival rates for advanced NSCLC, but only 20-30% of the patients treated with ICIs exhibit a long-term survival benefit. There is a lack of validated biomarkers and integrative multi-modal analyses to predict response and resistance.
Objective
The primary objective of QUANTIC is to develop and validate combined mechanistic mathematical models with machine learning, which are predictive of response and toxicity to ICI from baseline and early longitudinal data gathered from the PIONeeR project.
Data
QUANTIC relies on modeling unique, large scale (n = 450 patients), heterogeneous and longitudinal data collected during the PIONeeR (“Precision Immuno-Oncology for advanced Non-small cell lung cancer patients with PD-(L)1 ICI Resistance”) biomarkers clinical study.
Methodology
1. build mathematical models able to simulate the kinetics of response to treatments on the basis of biological hypotheses,
2. design adequate statistical methods able to identify parameters from the models using large-dimension data
3. establish patient-specific predictive models.
Funding
Inserm Plan Cancer MIC (Mathématiques et Informatique pour le Cancer), 2019 – 2022
Publications
Comprehensive biomarkers (BMs) analysis to predict efficacy of PD1/L1 immune checkpoint inhibitors (ICIs) in combination with chemotherapy: a subgroup analysis of the Precision Immuno-Oncology for advanced Non-Small CEll Lung CancER (PIONeeR) trial.
F. Barlesi, L. Greillier, …, J. Ciccolini, M. Karlsen, P. Dufossé,…, S. Benzekry
ESMO IO, Abstr 3MO, 2022
Comprehensive biomarkers analysis to explain resistances to PD1-L1 ICIs: The precision immuno-oncology for advanced non-small cell lung cancer (PIONeeR) trial.
L. Greillier, …, J. Ciccolini, M. Karlsen, P. Fleury, … S. Benzekry, J. Fieschi, F. Barlesi.
AACR, Abstr LB120, 2022
Deciphering the response to immune-checkpoint inhibitors in lung cancer with artificial intelligence-based analysis: the pioneer and quantic joint-projects.
J. Ciccolini, S. Benzekry, F. Barlesi.
British Journal of Cancer, 2020, 123 (3)
Updated news on the Pioneer Project
_________________________________________________________________________________________________________________________
Mechanistic modeling for the prediction of metastatic relapse in breast cancer
P.I.s : S. Benzekry, X. Muracciole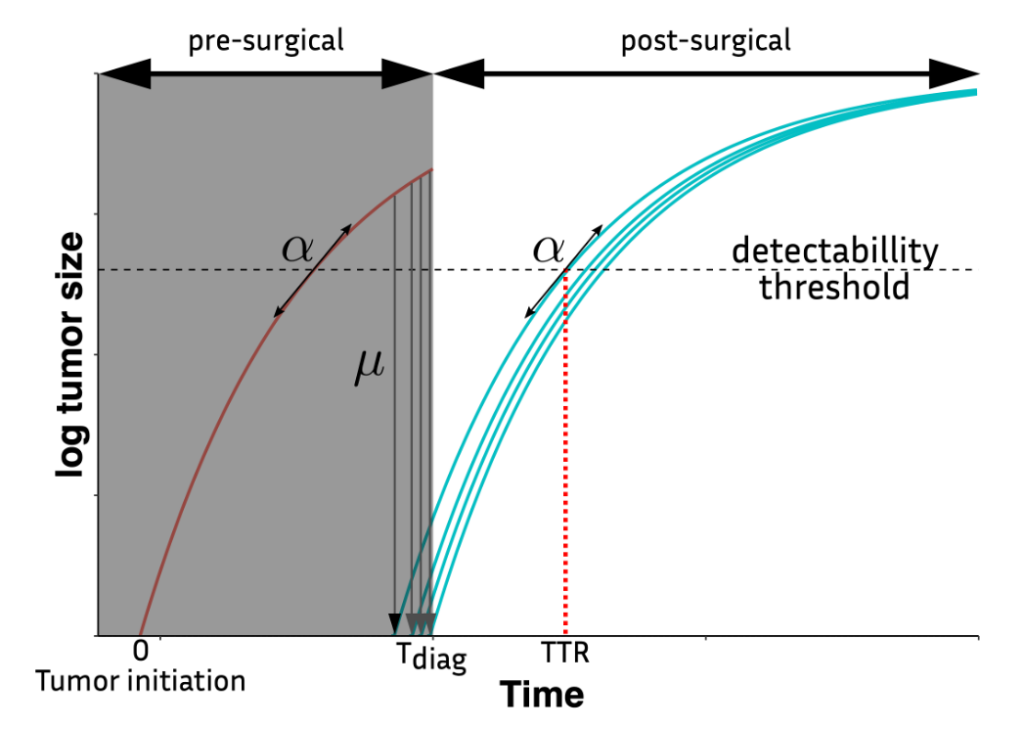
COMPO members involved : C. Bigarré, D. Barbolosi
Partners : G. MacGrogan (Bergonié Institute Bordeaux), F. Bertucci (Institut Paoli-Calmettes, CRCM)
Clinical need :
Estimation of the risk of metastatic relapse is a major challenge to decide treatment options for early-stage breast cancer patients.
Objective
The objective of this work is to use clinical data to calibrate a mechanistic model to predict metastatic relapse algorithms.
Data
We have access to clinical data from breast cancer patients from the Bergonié Institute (N = 1161 patients), the APHM radiotherapy department (N = 167) and several public databases with tumor genomic information aggregated by our partners at IPC (N = 992).
Methodology
To date, metastatic free survival (MFS) analysis mainly relies on classical – agnostic – statistical models (e.g., Cox regression). Instead, we propose to derive mechanistic models to predict MFS. We use the statistical framework of mixed-effects models to describe the variability of the parameters of the model in the population.
Publications:
Development and Validation of a Prediction Model of Overall Survival in High-Risk Neuroblastoma Using Mechanistic Modeling of Metastasis
S. Benzekry, C. Sentis, C. Coze, L. Tessonnier, N. André
JCO: Clin Cancer Informatics, Volume 5, pp. 81-90, medRxiv, 2021
Machine learning and mechanistic modeling for prediction of metastatic relapse in breast cancer
C. Nicolo, C. Perier, M. Prague, G. MacGrogan, O. Saut, S. Benzekry
JCO: Clinical Cancer Informatics, Volume 4, pp. 259-274, bioRxiv, 2020
Modeling spontaneous metastasis following surgery: an in vivo-in silico approach
S. Benzekry, A. Tracz, M. Mastri, R. Corbelli, D. Barbolosi, J.M.L. Ebos
Cancer Research, Volume 76, Issue 3, pp. 535-547, hal, 2016
_________________________________________________________________________________________________________________________
SChISM – Size CfDNA Immunotherapies Signature Monitoring
P.I.s : S. Benzekry, S. Salas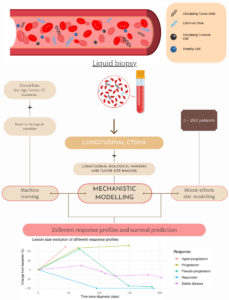
COMPO members involved : L. Nguyen Phuong
Partners : M. Lavielle (XPOP – Inria), Adelis, F. Fina (Adelis, ID-Solutions)
Clinical need
A major immuno-oncology issue is currently to early predict the response of
immunotherapy for cancer patients, ideally before the first imaging exam. Circulating DNA available from liquid biopsies are a promising, non-invasive, surrogate to imaging to early assess response during treatment.
Objective
The objective is to observe and model real-time variations in concentration and size of cell-free plasma DNA (cfDNA) to early predict response and survival following immunotherapy for five cancer types (melanoma, metastatic head and neck squamous-cell carcinoma, clear cell kidney cancer, urothelial bladder carcinoma and non-small cell lung cancer), using a novel, patented, standardized and innovative technology provided by our partners, ID-solutions and Adelis.
Data
We are collecting data on 260 patients starting an anti-PDL1/PD1 treatment (possibly adjuvant). These consist of baseline and longitudinal data, the latter beinglongitudinal data from standardized ID-Solutions/ADELIS extraction, corresponding to concentration and size of cfDNA fragments at different timepoints, biological (hematology and biochemistry) and imaging data.
Methodology
We are developing a mechanistic model to describe kinetics of cfDNA. Statistical nonlinear mixed-effects modeling is used to quantify inter-patient variability. Eventually, statistical machine learning methods are finally used to build predictive algorithms.
_________________________________________________________________________________________________________________________
Machine learning for prediction of response to immunotherapy using real world data
P.I.s S. Benzekry, L. Greiller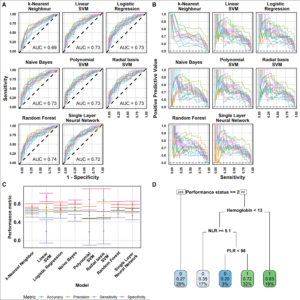
COMPO members involved D. Barbolosi, M. Karlsen, P. Fleury
Partners F. Barlesi (Gustave Roussy Institute)
Clinical need
Immune checkpoint inhibitors (ICIs) are now a therapeutic standard in advanced non-small cell lung cancer (NSCLC), but only 20% of patients do respond and strong predictive markers for efficacy are lacking.
Objective
Leverage clinical and biological data available from routine measurements to predict response and toxicity to ICIs using machine learning (ML) algorithms, in order to individually adapt therapeutic interventions.
Data
So far, we analyzed data of 298 second-line (or more) stage IV non-small cell lung cancer (NSCLC) patients, featuring 10 pre-treatment blood count variables together with clinical variables.
Methodology
We use data science techniques to study the association of these routine data with response, progression free survival and overall survival. The general supervised learning method consists of two main steps: feature selection and statistical learning using ML.
Publications:
Machine learning for prediction of immunotherapy efficacy in non-small cell lung cancer from simple clinical and biological data
S. Benzekry, M. Grangeon, M. Karlsen, M. Alexa, I. Bicalho-Frazeto, S. Chaleat, P. Tomasini, D. Barbolosi, F. Barlesi, L. Greillier
Cancers, Volume 5, pp. 81-90, medRxiv, 2021
Artificial intelligence and mechanistic modeling for clinical decision making in oncology
S. Benzekry
Clinical Pharmacology and Therapeutics, Volume 108, Issue 3, pp.471-486, 2020


