Associated-Team CytoDI — Quantitative Imaging of Cytoskeleton Dynamics in 3D
PRINCIPAL INVESTIGATORS
Charles KERVRANN, Research director, head of SERPICO Team-Project, INRIA Rennes
The SERPICO Team-Project, headed by Charles Kervrann, is a leading image processing group developing methodologies to decipher the spatiotemporal organization of intracellular structures. The team investigates non-parametric statistical methods, robust statistics, stochastic filtering and image representation to push the envelope in image denoising, segmentation and motion estimation and to tackle challenges in live cell imaging.
Gaudenz DANUSER, Professor of Cell Biology, UT SouthWestern Medical Center, Dallas, USA
The Danuser Lab is a leading quantitative cell biology lab focusing on mechano-chemical signaling involved in cellular morphogenesis. The lab investigates multi-dimensional live cell imaging combined with statistical modeling for a minimally invasive study of cell functions. The Danuser Lab, now at UT Southwestern Medical center, is moving closer to clinical research to study cell morphogenesis in oncogenic pathways.
ABSTRACT
The main scientific goal of the Associated-Team (AT) CytoDI is the spatiotemporal characterization and comparison of cytoskeleton networks involved in cell migration and observed through high-resolution live cell imaging in three dimensions (3D). Cell migration is one of the best-studied phenomena in biology and medicine since the underlying mechanisms are known to facilitate the dissemination of tumor cells in blood and organs and eventually the formation of secondary tumors and metastases.
In fundamental cell biology, it has been established that the cytoskeleton, composed of actin filaments, microtubules and intermediate filaments is essential for cell migration:
Nevertheless, the mechanical and chemical states of migrating cells under various external conditions remain largely unknown. At present, the main challenge is to understand how a 3D motile cell crawls through the 3D extracellular matrix.
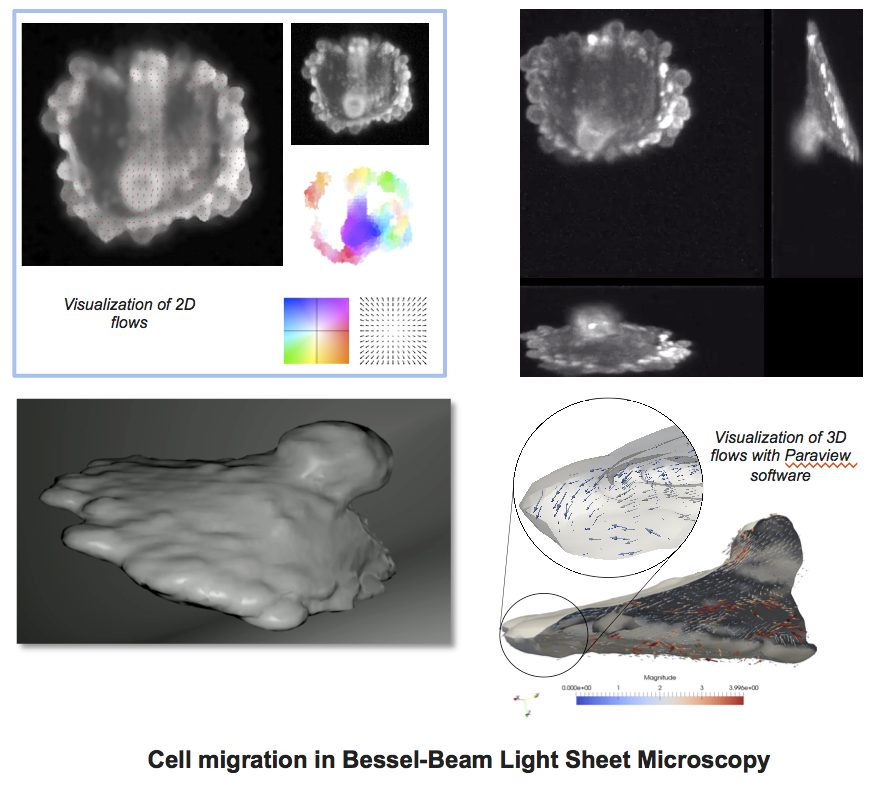
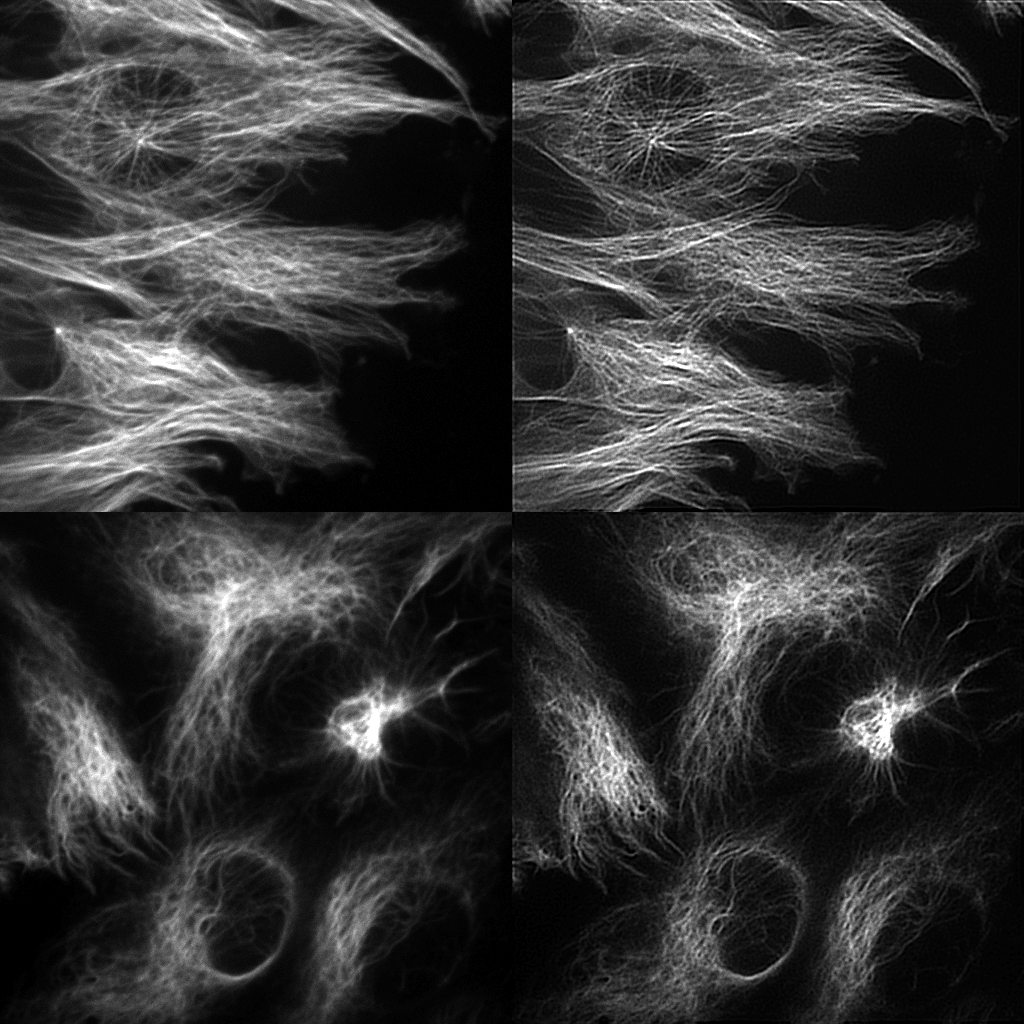
Since a few years, we have the opportunity to observe the MT and IF in 3D + time with Bessel beam light-sheet microscopy (LSM) (Planchon et al. 2011), (Dean et al. 2015). In LSM, cells can be typically observed deeply within organotypic tissue models that provide a realistic biological context. Nevertheless, the corresponding microscopy data on spatiotemporal distributions of observed dynamics are semi quantitative at best. Accordingly, the proposed AT CytoDI targets the development of high-end signal processing and computer vision tools to unfold the dynamical coordination of MT and IF networks in 3D. The objective for the 3 years is to design local and global descriptors of the spatial conformation, motion and deformation of the cytoskeleton. One important issue is the development of 3D optical flow methods to quantify dense motion fields of filaments. Another issue is the extraction of filaments for image registration and comparison. This includes image deconvolution (see figure). In this area, general metrics to compare and classify the cytoskeleton components are currently investigated. All these algorithms will be designed to extract and represent dynamics information and motion for a very active research field in cell biology and imaging. The experiments will be carried out on oncogenically transformed lung cancer epithelial cells.
Keywords:image analysis, motion analysis, metrics for comparison, fluorescence microscopy, cell migration, cytoskeleton, microtubules, intermediate filaments, cancer.
MOTIVATION
The mechanical and chemical states of migrating cells under various external conditions in the 3D extracellular matrix remain largely unknown.
MAIN OBJECTIVE
Development of high-end signal processing and computer vision tools to unfold the dynamical coordination of MT and IF networks in 3D.
-
Development of3D optical flow methods to quantify dense motion fields of filaments.
-
Extraction of filaments for comparison.
-
Experiments on oncogenically transformed lung cancer epithelial cells.
PROJECT OVERVIEW
The main scientific goal is the spatiotemporal characterization and comparison of cytoskeleton networks observed through live cell imaging in 2D and 3D. We address the five following tasks:
- Task 0: Preparation of biological samples and experiments in cell biology, image acquisition in high-resolution microscopy (LSM).
- Task 1: Design of 3D dense optical flow in LSM and feature tracking algorithms to provide local descriptors of network dynamics.
- Task 2: Extraction of cytoskeleton structure and filament networks.
- Task 3: Analysis of motion vector fields and sparse descriptors to compute rigidity and relevant biophysical features of the cytoskeleton (e.g. stiffness and filament curvature).
- Task 4: Analysis of metrics to compare MT and IF network dynamics and organization in migrating and metastatic cells in a physiological environment.
One important benefit of the collaboration is the ability to simultaneously tune the parameters of data acquisition and the design of algorithms to obtain high precision in measuring the biological phenotypes.
MEETINGS
Meeting in Dallas (May 18-20, 2016, USA, TX) (Visit of C. Kervrann, P. Bouthemy (Serpico Team)): The first meeting of the AT CytoDI took place in Dallas in May 2016 as originally scheduled, to discuss and update current research direction and discuss scientific progress. The different solutions for 3D microscopy developed in the Danuser lab by R. Fiolka and K. Dean were presented to lay out the potential of newly developed setups such as acquisition speed, resolution and signal-to-noise ratio trade-off. A general presentation of both team on-going project where presented, underlining in particular the current frontier met by the Danuser lab in the quantification of 3D imaging data. Finally, an in-depth discussion between the Serpico team and the cytoskeleton team in the Danuser lab (Z. Gan and T. Zhang) laid out recent challenges brought by the filament interaction measurement in both 2D and 3D, such as the current impossibility to measure the interaction between the protein PKC and Vimentin disassembly, or observing the annealing of vimentin filament along http://www.gulfportpharmacy.com microtubule fibers due to limited detection power of the steerable-filter-based filament detection technique.
Meeting in College Station (January 4-7, 2017, USA, TX) (C. Kervrann & V. Briane (PhD student) (Serpico Team) and P. Roudot (Danuser lab)): The second meeting of the AT CytoDI took place in College Station in January 2017 to discuss scientific progress. The next steps and decisions are the following ones:
- Deconvolution of filaments is promising and new experiments will be performed.
- A first prototype of 3D optical flow algorithm based on block matching techniques will be developped in the fortcoming weeks by Sandeep Manandhar.
- A Master student will be recruited in March 2017 to develop a statistical method for detecting the local and global co-orientation of two filament networks.
- A visit of P. Roudot and K. Dean (Danuser lab) is planned in Rennes (June 2017).
Meeting in Rennes (July 17-21, 2017): The second meeting of the AT CytoDI took place in Rennes in July 2017 (visit of P. Roudot and K. Dean), to discuss and update current research direction and discuss scientific progress. A one-day visit at Curie Institute was organized to present the Lattice Light Sheet Microscopy set-up, which is very close to the 3D microscopy solutions developed in the Danuser lab. A meeting between the collaboration members, Dhiraj Bhatia and Cesar Augusto Valades Cruz from the Johannes Lab discussed the quantification challenges posed by the study of endocytic vesicle dynamic in 3D. Several meetings were organized with students (S. Manandhar, V. Briane, E. Moebel, T. Dubois, Q. Delannoy) to synchronize development in optical flow, co-orientation
and visualization. The Danuser team focused on presenting recent imaging and analysis capacities as well as the current solution in development for the systematic analysis, contextualization and interpretation of 3D dynamics for quantitative biology.
Visit of Sandeep Manandhar (PhD student) at UTSW Dallas, March 1-31, 2018: The objective of this visit was for Sandeep Manandhar to i) get a deeper understanding of bioimaging capacities and limitations in the context of biological studies, 2) to refine the design and validation of his approaches in this context and 3) returning from his visit with a clear vision for biologically relevant tools. For the Danuser lab, the objective was 1) to evaluate the breakpoint of path-match-based approach toward heterogeneous motion 2) to fill the gap between object tracking and generic motion estimation for studies such as actin speckle or collagen.
Meeting in Rennes (January 8-11, 2019) with C. Kervrann, P. Bouthemy, and S. Manandhar (PhD student) (Serpico team) and P. Roudot (Danuser lab)
SEMINARS
- C. Kervrann, May 19th, 2016, Seminar UTSW, Danuser lab, Dallas: Computational analysis of intracellular membrane dynamics: from live cell images to biophysical model.
- P. Roudot, July 17th, 2017, Seminar Inria Serpico Team, Rennes: Piecewise-stationary motion modeling and iterative smoothing to track heterogeneous motion in dense intracellular environments.
- S. Manandhar, March, 2018, Seminar UTSW, Danuser’s lab, Dallas: A sparse-to-dense method for 3D optical flow estimation in 3D light-microscopy image sequences.
JOINT PARTICIPATIONS TO EVENTS AND WORKSHOPS
- Co-organization (P. Bouthemy and C. Kervrann, Serpico Team) of a mini-symposium (Image Analysis Advances in Dynamic Microscopy and Live Cell Imaging / Invited speakers : G. Danuser & P. Roudot (Danuser lab)) at the SIAM Imaging Sciences conference, 2016, Albuquerque, New-Mexico, USA.
- Participation to the international Quantitative BioImaging (QBI 2017) conference, C. Kervrann & V. Briane (Serpico Team), P. Roudot & K Dean (Danuser lab), College Station, USA, TX.
- Participation to the international NEUBIAS 2018 conference, Szeged, Hungary, January 31 – February 2, 2018, C. Kervrann and G. Danuser.
- Participation to the international “Quantitative BioImaging” (QBI 2019) conference, Rennes.
SUPERVISION OF STUDENTS AND ENGINEERS
- A. Diouf (Master, INSA Rouen, 4 months, 2016): Proximal methods for image deconvolution in fluorescence microscopy: application of microtubule and vimentin filament images.
- A. Caranfil (Engineer, 2016): Evaluation of local and global methods for 3D optical flow estimation in diaSLM and ASLM microscopy.
- S. Mandahar (Phd student, since October 2016): 3D Optical flow computation in microscopy.
- Q. Delannoy (Internship, Master 1, ENSAI, 2-3 months, 2017): Statistical analysis of coorientation of filament structures in 2D fluorescence microscopy.
- T. Dubois (Internship, Master 1, ENSAI 2-3 months, 2017): Statistical analysis of co-direction of motion fields in 2D fluorescence microscopy.
JOINT PUBLICATIONS
- Roudot, P., Ding, L., Buckhart, C., Jaqaman, K., Kervrann, C. and Danuser, G. Tracking heterogeneous motion in dense condition. IEEE Trans. Image Processing, 26(11) : 5395-5410, 2017.
- Manandhar, S., Bouthemy, P., Welf, E., Roudot, P. and Kervrann, C. A sparse-to-dense method for 3D optical flow estimation in 3D light-microscopy image sequences. In Proc. IEEE International Symposium on Biomedical Imaging (ISBI 2018), 952–56, 2018.
- Manandhar, S., Bouthemy, P., Welf, E., Roudot, P. and Kervrann, C. 3D flow field estimation and assessment for live cell fluorescence microscopy. Bioinoformatics 2019.