DenseMapping software: Dense mapping of diffusion and drift of moving particules
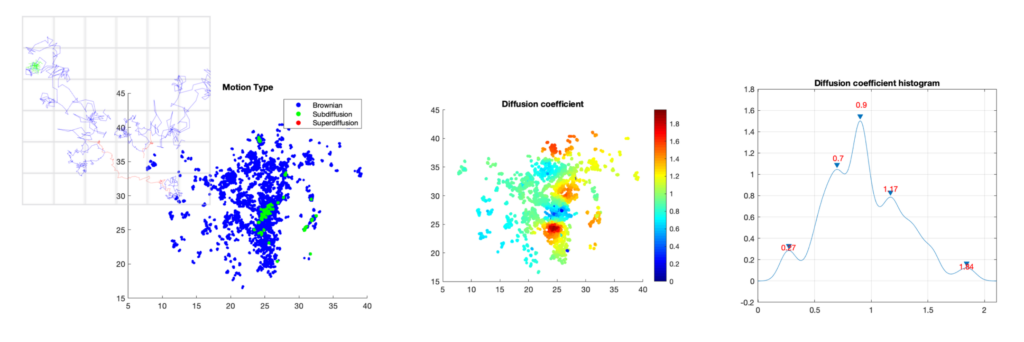
Analysis of dynamics of trancription factors in the nucleus. Left: classification of trajectories into three diffusion modes: sub-diffusion (green), super-diffusion (red), and Brownian motion (blue); middle: 2D map of the estimated diffusion coefficient; right: histogram of the 2D map.
Overview
It is of primary interest for biologists to be able to visualize the dynamics of proteins within the cell. DenseMapping software allows to estimate the diffusion and drift parameters attached to moving biomolecules within cells from 2D/3D individual trajectories. In a first step, each particle track is labeled into three motion categories: confined motion (subdiffusion), Brownian motion (free diffusion), and directed motion (superdiffusion). The long trajectories are also segmented into sub-trajectories according to these three categories. In a second step, two 2D/3D maps (diffusion, drift) are computed from a local analysis of the trajectories. The local spatio-temporal kernel estimators correspond to weighted averages of the trajectory elements. The weights allow to select and aggregate the information in an optimal way.
Software distribution
Work in progress
References
- A. Salomon, C. Valades-Cruz, L. Leconte, C. Kervrann. Dense mapping of intracellular diffusion and drift from single particle tracking data analysis, Proc IEEE Int. Conf. Acoustics, Speech and Signal Processing (ICASSP’20), Barcelona, Spain, May, 2020
- V. Briane, C. Kervrann, M. Vimond. Statistical analysis of particle trajectories in living cells, Phys. Rev. E 97, 062121 (doi:10.1103/PhysRevE.97.062121), 2018 (arXiv) (HAL-INRIA-01557705)
- V. Briane, M. Vimond, C. Valades Cruz, A. Salomon, C. Wunder, C. Kervrann. A sequential algorithm to detect diffusion switching along intracellular particle trajectories, Bioinformatics, btz489, HAL-INRIA-01966831, 2019
- V. Briane, M. Vimond, C. Kervrann. An overview of diffusion models for intracellular dynamics analysis, bbz052, Briefings in Bioinformatics, 21(4): 1136–1150, HAL-INRIA-0196682, 2020