Bacteria provide fascinating examples of the capacity of microorganisms to adapt to changes in their environment. The adaptive responses of bacteria are controlled by large and complex regulatory networks that involve genes, mRNAs, proteins, small effector molecules, and metabolites. The study of bacterial regulatory networks requires experimental tools for characterizing the interactions making up the networks and measuring the dynamics of cellular processes on the molecular level. In addition, when dealing with systems of this size and complexity, we need mathematical modelling and computer simulation to integrate available biological data, and understand and predict the dynamics of the system under various environmental and physiological conditions. The analysis of living systems through the combined application of experimental and computational methods has gathered momentum in recent years under the name of systems biology.
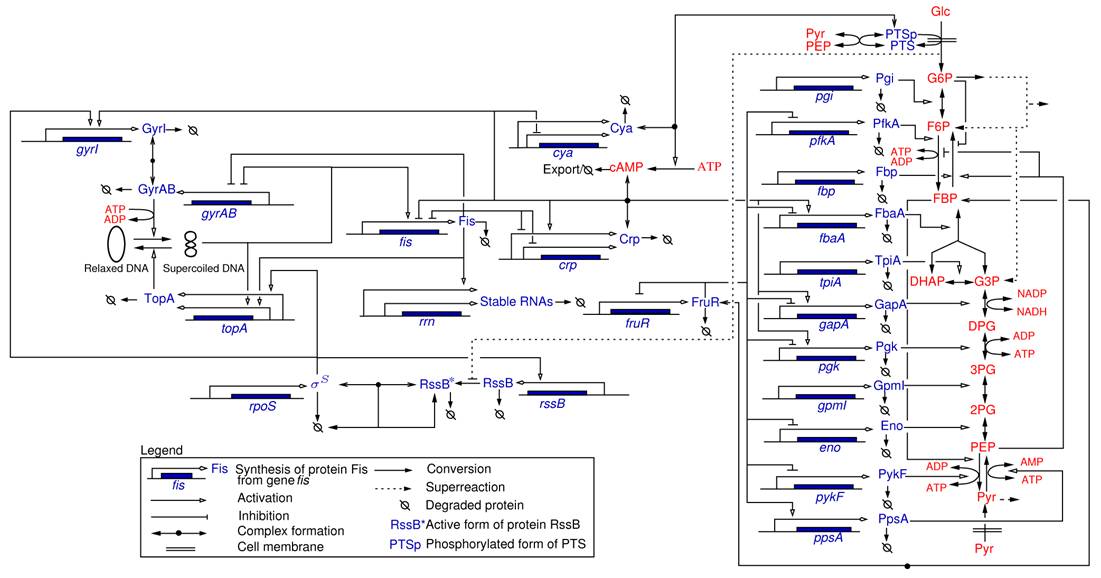
Regulatory network in the enterobacterium E. coli (Baldazzi et al., PLoS Comput. Biol., 2010)
The first aim of the IBIS project-team is the unravelling of the adaptive strategies of bacteria through a systems-biology approach, making use of both models and experiments. In particular, we will focus on the enterobacterium Escherichia coli, for which enormous amounts of genomic, genetic, biochemical and physiological data have been accumulated over the past decades. A better understanding of the adaptive capabilities of E. coli in a variety of situations is a necessary prerequisite for interfering with these strategies by specific perturbations or by even rewiring the underlying regulatory networks. This is the second and most ambitious aim of the research programme, which does not only spawn fundamental research on the control of living matter, but which may ultimately acquire practical relevance since E. coli serves as a model for many pathogenic bacteria and is widely used in biotechnology.
The aims of IBIS raise four main challenges that generate new problems on the interface of (experimental) biology, applied mathematics, and computer science. In particular, the research program of the project-team focuses on
- the modeling and analysis of integrated metabolic and gene regulatory networks;
- the inference of regulatory networks from time-series data;
- natural and engineered control of regulatory networks;
- the analysis of the qualitative dynamics of gene regulatory networks.
IBIS joins researchers from INRIA Grenoble – Rhône-Alpes and the Laboratoire Interdisciplinaire de Physique at Université Grenoble Alpes (CNRS UMR 5588).
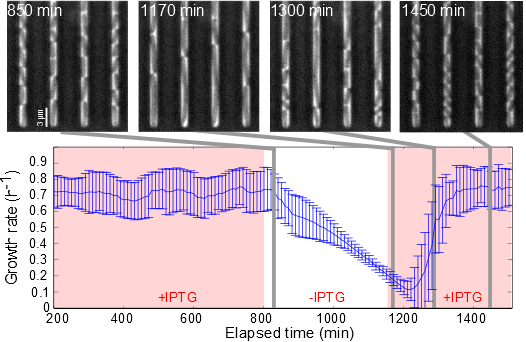
External growth-rate control of an E. coli strain in a microfluidics device (Izard, Gomez Baldaras et al., Mol. Syst. Biol., 2015)


