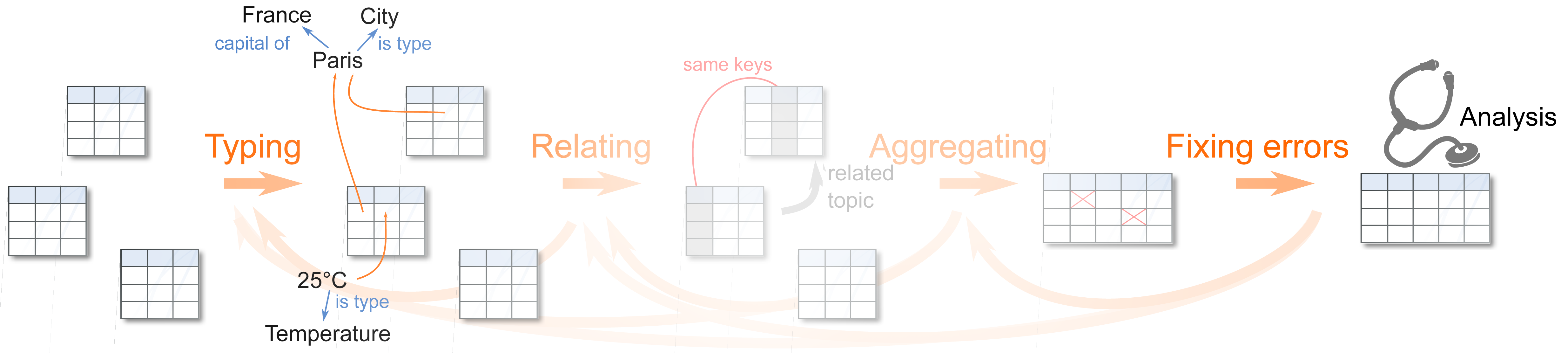
Representation learning for heterogeneous databases
We develop a deep-learning methodology on heterogeneous databases, comprising relational data and tabular datasets at large scale.
The stakes are (1) to build machine-learning models that apply readily to the raw, uncurated data so as to avoid manual cleaning, data formatting and integration, (2) to extract reusable representations that reduce sample complexity on new databases by transforming the data in well-distributed vectors.
The challenges are:
- Tokens (symbolic entries) often with high cardinality, such ICD10 classification of disease
- Numerical entries with different, non-Gaussian marginals, such as salary (long-tailed distribution) or age
- Missing values, often missing not at random and thus not ignorable without causing biases
- Shoft text (possibly corresponding to non-normalized entities)
- Operation across tables: aggregation (learning on sequences, joins (optimal transport)
These challenges call for dedicated architectures, that borrow from neural architectures used in natural language
processing and add new tricks. The fundamental ingredients are:
– Entity embeddings in tabular and relational data
– Neural-network formulations of classical statistical techniques such General Additive Models or EM for missing values (Le Morvan et al., 2020);
– Simple attention and self-attention mechanisms, as those we developed for short texts (Chen et al., 2021);
Current research focus
- Learning despite database normalization errors
- Tabular deep learning
Publications
- titre
- On the consistency of supervised learning with missing values
- auteur
- Julie Josse, Jacob M. Chen, Nicolas Prost, Erwan Scornet, Gaël Varoquaux
- article
- 2024
- Accès au texte intégral et bibtex
-


- titre
- Causal inference methods for combining randomized trials and observational studies: a review
- auteur
- Bénédicte Colnet, Imke Mayer, Guanhua Chen, Awa Dieng, Ruohong Li, Gaël Varoquaux, Jean-Philippe Vert, Julie Josse, Shu Yang
- article
- Statistical Science, In press
- Accès au texte intégral et bibtex
-


- titre
- Evaluating machine learning models and their diagnostic value
- auteur
- Gaël Varoquaux, Olivier Colliot
- article
- Olivier Colliot. Machine Learning for Brain Disorders, Springer, 2023
- Accès au texte intégral et bibtex
-


- titre
- Relational Data Embeddings for Feature Enrichment with Background Information
- auteur
- Alexis Cvetkov-Iliev, Alexandre Allauzen, Gaël Varoquaux
- article
- Machine Learning, 2023, 112 (2), pp.687-720. ⟨10.1007/s10994-022-06277-7⟩
- Accès au texte intégral et bibtex
-


- titre
- Machine learning for medical imaging: methodological failures and recommendations for the future
- auteur
- Gaël Varoquaux, Veronika Cheplygina
- article
- npj Digital Medicine, 2022, 5 (1), pp.48. ⟨10.1038/s41746-022-00592-y⟩
- Accès au bibtex
-


- titre
- Causal effect on a target population: a sensitivity analysis to handle missing covariates
- auteur
- Bénédicte Colnet, Julie Josse, Gaël Varoquaux, Erwan Scornet
- article
- Journal of Causal Inference, 2022, 10 (1), pp.372-414. ⟨10.1515/jci-2021-0059⟩
- Accès au texte intégral et bibtex
-


- titre
- How to remove or control confounds in predictive models, with applications to brain biomarkers
- auteur
- Darya Chyzhyk, Gaël Varoquaux, Michael Milham, Bertrand Thirion
- article
- GigaScience, 2022, 11, ⟨10.1093/gigascience/giac014⟩
- Accès au texte intégral et bibtex
-


- titre
- Analytics on Non-Normalized Data Sources: more Learning, rather than more Cleaning
- auteur
- Alexis Cvetkov-Iliev, Alexandre Allauzen, Gaël Varoquaux
- article
- IEEE Access, In press, 10, pp.42420-42431. ⟨10.1109/ACCESS.2022.3168013⟩
- Accès au texte intégral et bibtex
-


- titre
- Benchmarking missing-values approaches for predictive models on health databases
- auteur
- Alexandre Perez-Lebel, Gaël Varoquaux, Marine Le Morvan, Julie Josse, Jean-Baptiste Poline
- article
- GigaScience, In press, ⟨10.1093/gigascience/giac013⟩
- Accès au texte intégral et bibtex
-


- titre
- AI as statistical methods for imperfect theories
- auteur
- Gaël Varoquaux
- article
- NeurIPS 2021 – 35th Conference on Neural Information Processing Systems. Workshop: AI for Science, Dec 2021, Virtual, France
- Accès au texte intégral et bibtex
-


- titre
- What’s a good imputation to predict with missing values?
- auteur
- Marine Le Morvan, Julie Josse, Erwan Scornet, Gaël Varoquaux
- article
- NeurIPS 2021 – 35th Conference on Neural Information Processing Systems, Dec 2021, Virtual, France
- Accès au texte intégral et bibtex
-


- titre
- Accounting for variance in machine learning benchmarks
- auteur
- Xavier Bouthillier, Pierre Delaunay, Mirko Bronzi, Assya Trofimov, Brennan Nichyporuk, Justin Szeto, Naz Sepah, Edward Raff, Kanika Madan, Vikram Voleti, Samira Ebrahimi Kahou, Vincent Michalski, Dmitriy Serdyuk, Tal Arbel, Chris Pal, Gaël Varoquaux, Pascal Vincent
- article
- MLsys 2021 – 4th Conference on Machine Learning and Systems, Apr 2021, San Francisco (virtual), United States
- Accès au texte intégral et bibtex
-


- titre
- A lightweight neural model for biomedical entity linking
- auteur
- Lihu Chen, Gaël Varoquaux, Fabian Suchanek
- article
- AAAI 2021 – The Thirty-Fifth Conference on Artificial Intelligence, Association for the Advancement of Artificial Intelligence, Feb 2021, Palo Alto (virtual), United States. pp.12657-12665
- Accès au texte intégral et bibtex
-


- titre
- Preventing dataset shift from breaking machine-learning biomarkers
- auteur
- Jérôme Dockès, Gaël Varoquaux, Jean-Baptiste Poline
- article
- GigaScience, In press, ⟨10.1093/gigascience/giab055⟩
- Accès au texte intégral et bibtex
-


- titre
- NeuMiss networks: differentiable programming for supervised learning with missing values
- auteur
- Marine Le Morvan, Julie Josse, Thomas Moreau, Erwan Scornet, Gaël Varoquaux
- article
- NeurIPS 2020 – 34th Conference on Neural Information Processing Systems, Dec 2020, Vancouver / Virtual, Canada
- Accès au texte intégral et bibtex
-