GenLoc3D: A Fiji plugin for 3D gene localization in live cells
Overview
GenLoc3D plugin (version 1.0 – november 2015) implements a 3D high-precision gene localization algorithm in cell nuclei [1] (Please cite [1] if you use this plugin for your work.)
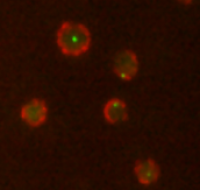
This software computes the distance between the gene and the nuclear periphery observed in two 16 bits 3D TIFF stacks. Tagged gene (usually marked in green) and tagged nucleoporins (usually marked in red) are observed in those two stacks (as in Fig. 1). The output consists in two 3D TIFF images, for the detected genes and nucleus periphery, and an Excel Table with the computed distances and radius of gene with respect to the nucleus periphery in each cell. Gene/nuclear periphery distances can be compared between two acquisition conditions on the same cell field.
Software distribution
The downloaded file consists in a JAR executable file. This version is compatible with Linux (64 bit), Windows 7 (64 bit) and MacOX. Please, contact giovanni.petrazzuoli@inserm.fr if you have any problem of compatibility with your operating system.
- Download GenLoc3D_.jar
- Download the GenLoc3D documentation
This plugin has been written and developed by Suman Maji*, Giovanni Petrazzuoli*, Charles Kervrann, David Guet*, Laura T. Burns, Jerome Boulanger, Pascal Hersen, Susan R. Wente, Jean Salamero, Catherine Dargemont* / * University Paris Diderot, Sorbonne Paris Cité, INSERM U944, CNRS UMR7212, Hopital Saint Louis, Paris, France
Fundings
- LABEX WAI (grant ANR-11-LABX-0071)
- Fondation pour la recherche medicale
Reference
[1] D. Guet, L.T. Burns, S. Maji, J. Boulanger, P. Hersen, S.R. Wente, J. Salamero and C. Dargemont. Combining Spinach-tagged RNA and gene localization to image gene expression in live yeast – Nature Communications| 6:8882 | DOI: 10.1038/ncomms9882, 2015 (see http://www.nature.com/ncomms/2015/151119/ncomms9882/abs/ncomms9882.html)