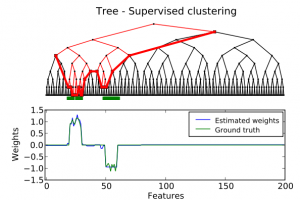
Reverse inference, or brain reading, is a recent paradigm for analyzing functional magnetic resonance imaging (fMRI) data, based on pattern recognition and statistical learning. This approach aims at decoding brain activity by predicting some cognitive variables related to brain activation maps. Reverse inference takes into account the multivariate information between voxels and is currently the only way to assess how precisely some cognitive information is encoded by the activity of neural populations within the whole brain. However, it relies on a prediction function that is plagued by the curse of dimensionality, since there are far more features than samples, i.e., more voxels than fMRI volumes. To address this problem, different methods have been proposed, such as, among others, univariate feature selection, feature agglomeration and regularization techniques. In this work, we consider a sparse hierarchical structured regularization. Specifically, the penalization we use is constructed from a tree that is obtained by spatially-constrained agglomerative clustering. This approach encodes the spatial structure of the data at different scales into the regularization, which makes the overall prediction procedure more robust to inter-subject variability. The regularization used induces the selection of spatially coherent predictive brain regions simultaneously at different scales. We test our algorithm on real data acquired to study the mental representation of objects, and we show that the proposed algorithm not only delineates meaningful brain regions but yields as well better prediction accuracy than reference methods.
-
News & Events
Research Projects
Meta