Population imaging relates features of brain images to rich descriptions of the subjects such as behavioral and clinical assessments. We use predictive analysis pipelines to extract functional biomarkers of brain disorders from large-scale datasets of resting-state functional Magnetic Resonance Imaging (R-fMRI), diffusion Magnetirc Resonance Imaging (dMRI), Magnetoencephalography (MEG) and Electroencephalography (EEG). We also built tools for automated data analysis which facilitate processing large datasets at scale. Some of our results are highlighted below.
Construction of surrogate biomarkers from brain images and electrophysiological signals
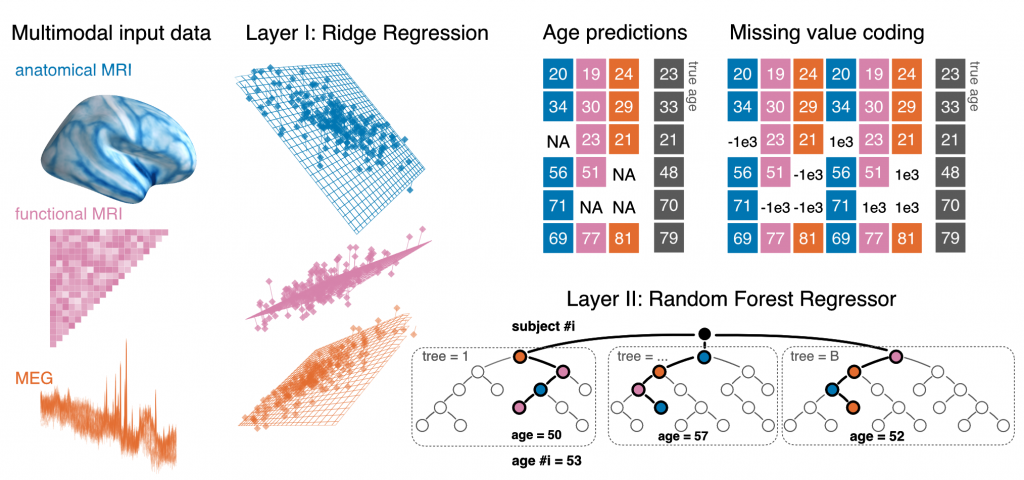
Electrophysiological methods, such as M/EEG and imaging methods (MRI, fMRI) provide unique views into brain health. Yet, when building predictive models from brain data, it is often unclear how multiple neuroimaging methods should be combined. Information can be redundant, useful common representations of multimodal data may not be obvious and multimodal data collection can be medically contraindicated, which reduces applicability. We propose to build surrogate biomarkers by approximating commonly available outcomes from multiple imaging modalities to then extract derived predictors for statistical modeling in clinical situations. A common approximation target choice for a surrogate biomarker is the age of a person. When approximating age from brain signals using machine learning, the resulting prediction errors systematically reflect individual aging and neurodegenerative processes, hence, the term brain age. Multimodal models of brain age can be conveniently built using the stacking method.
Our latest multimodal brain age model combining MEG, fMRI and MRI has been published in eLife.
Large-scale predictive modeling with M/EEG
M/EEG provides a unique window on brain function as it captures non-invasively neuronal large-scale dynamics in real-time across multiple time scales from seconds to less than milli-seconds. However, it was only recently that the availability of large datasets has rendered M/EEG an option for population-level predictive modeling in clinical neuroscience research. While some important steps of preprocessing can nowadays be automated, cross-person, cross-protocol and cross-site learning introduce peculiar challenges related to domain-adaptation problems and hierarchical variance components.n this research project we develop methods to improve learning across subjects with heterogenous data and conduct applied research in areas such as biomarker development and automation of medical diagnosis.
A framework for regression modeling from M/EEG signals
Linking biomedical outcomes to brain signals revealed by M/EEG comes with certain challenges as the brain sources are not directly observed inside the brain but outside of the brain with sensors capturing electromagnetic fields induced by the brain sources. To build appropriate regression models, it is essential to consider simultaneously separate generative models of the MEG signals (how are the M/EEG measurements related to brain sources?) and the the biomedical outcome (how is the outcome, e.g., age related to the brain sources?). Depending on the situation common regression techniques based on linear models can be directly applied on the sensor space signals (fully linear generative model) or additional modeling steps may be needed to take into account the electromagnetic field-spread (non-linear generative model for the outcome).
For theoretical analysis and empirical benchmarks see our paper published in NeuroImage.
Manifold regression to predict from M/EEG in the absence of source localization
Brain rhythms are a key source of information in electrophysiological modeling. Yet, they are highly exposed to geometric distortions when learning from non-invasive recordings. Closing this gap typically requires biophysical source modeling, which depends on availability of MRI recordings and specialized human expertise. In this paper we provide consistency proofs for appropriate generative models for learning from brain rhythms. We demonstrate with empirical M/EEG data that the consistent regression models also turn out more robust in the light of model violations induced by the cross-subject learning setting.
This paper is supported by the joint Inserm-Inria 2018 project and has been presented at NeurIPS 2019 (main track) and won the JDSE best paper paper award.
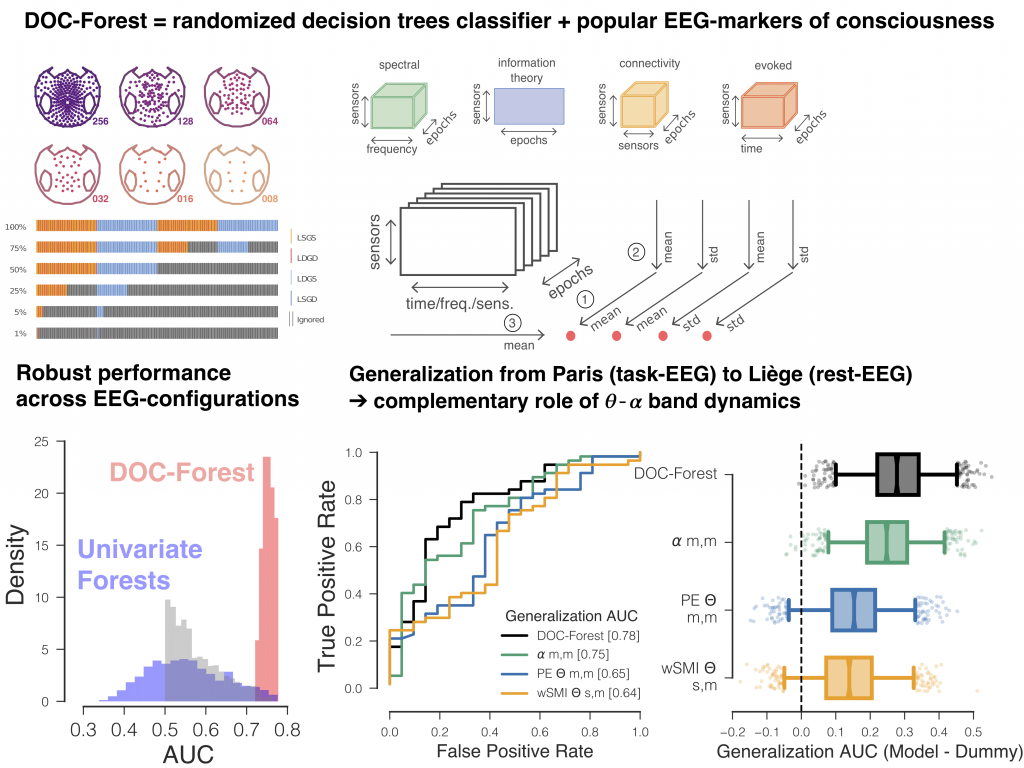
Robust cross-site and cross-protocol classification of EEG-based diagnosis in disorders of Consciousness
Diagnosis in severely brain injured patients is notoriously hard and heterogeneity of clinical data poses severe challenges for machine learning approaches to gain traction. In this work we demonstrate that combining multiple state-of-the-art EEG-markers of consciousness together with robust tree-based classification methods enable out-of-the box generalization between data from different EEG-protocols and hospitals.
This paper has been published in Brain together with a commentary article by Sokoliuk & Cruse discussing the implication of our findings in terms of neurophysiology of consciousness.
Joint prediction of multiple scores captures better individual traits from brain images
Benchmarking Functional Connectivity models:
-
An Autism-based example and a cross-dataset study
-
Imaging-psychatry challenge (IMPAC): predicting autism
Autoreject: Automated artifact rejection for MEG and EEG data
Removing artifacts from EEG and MEG signals is a common and necessary step in data analysis and, unfortunately, has claimed significant investment of human attention in the past. I developed and evaluated a novel algorithm, termed autoreject, for detecting and handling contaminated MEG and EEG data segments. Autoreject is described in Jas et al 2017 and is readily usable in a “plug and play” manner in a wide array of situations and has been validated on more than 250 datasets featuring a reanalysis of the Human Connectome Project MEG data. Notably, its successful usage does not require deep understanding of the method as it uses machine learning technology to handle artifact rejection in a data-driven manner, hence, reducing human processing time. It will soon be disseminated through the MNE Software. The code is accessible on github.
Statistical challenges in neuroimaging group-level analysis
Functional connectivity analysis: spatial patterns and covariance models
Charting the brain: Joint modeling of anatomical and functional brain features.
Meta-analysis of neuroimaging studies
Controlling a confound in predictive models
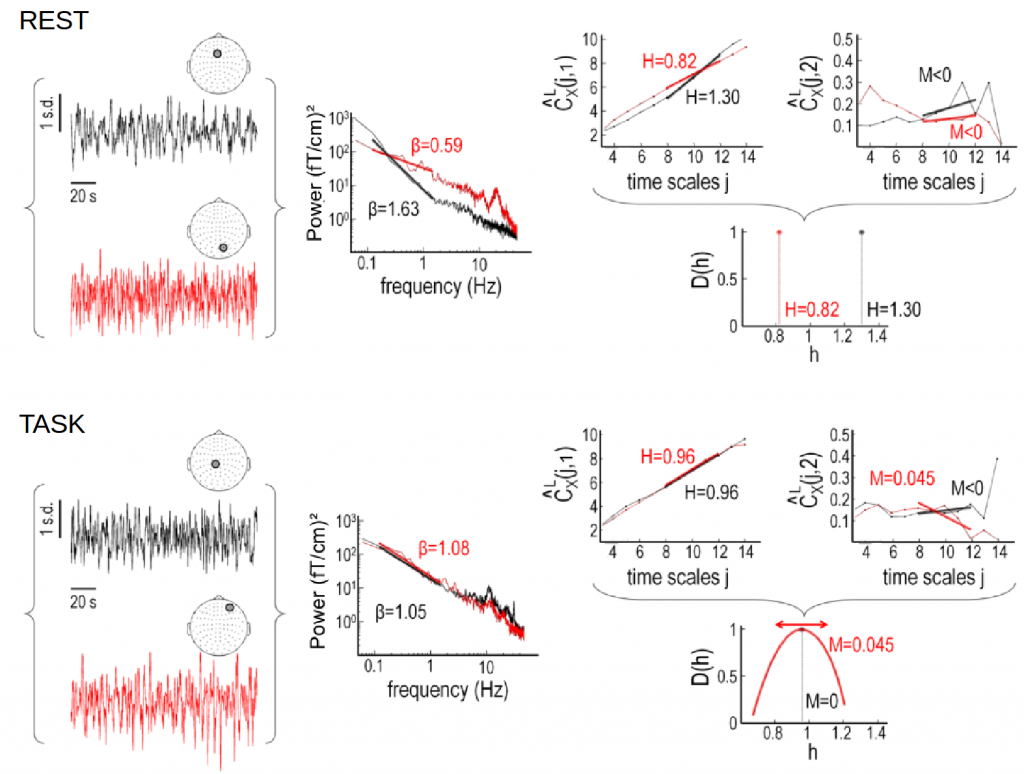
Self-similarity and multifractality in MEG
Here we investigate scale-free dynamics in brain activity. The temporal structure of macroscopic brain activity displays both oscillatory and scale-free dynamics. While the functional relevance of neural oscillations has been largely investigated, both the nature and the role of scale-free dynamics in brain processing have been disputed. Relying on the wavelet-leader multifractal formalism, we estimated self-similarity and multifractal exponents from resting-state and task MEG recordings.
Factors of mental health: an imaging approach
NeuroLang: Querying Heterogeneous Neuroimaging Databases and Probabilistic Representation Neuroanatomical Knowledge
NeuroLang is a probabilistic programming language for knowledge representation based on Datalog. We develop a query language which allows the user to write queries as a simple program in a straightforward non-composite syntax. NeuroLang merges heterogeneous datasets and their analysis with a near-English textual syntax using first order logic. It is a key property of the language to be intuitive for researchers outside computer science or not habituated to high-level programming. A general form of recursion allows expression of complex relationships from combining many datasets.
These methods enable NeuroLang to map cortical neuroanatomy by formally describing sulcal relationships, and for the incremental identification of cortical landmarks in a top-down order intuitive to neuroanatomists. NeuroLang allows the user to map subject-specific cortical landmarks with sulcus-specific queries. The primary sulci are gold-standard cortical landmarks and form the starting blueprint. From these, lower-level sulci can be identified from relations to the primaries and sulcal characteristics embedded into the language as predicates.
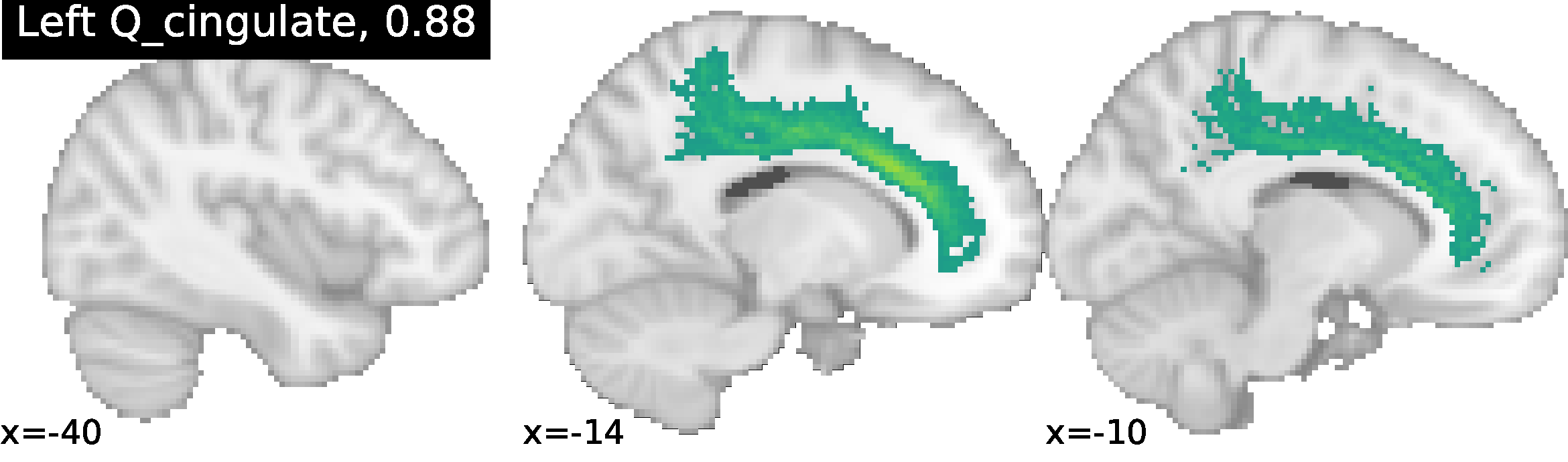
Identification of the Cingulate sulcus using NeuroLang queries, with the ratio of correct identification in 52 subjects. More examples can be found at https://hal.archives-ouvertes.fr/hal-02879734/document.
Brain tissue microstructure using Diffusion MRI
Non-invasive imaging at the cellular level could lead us to quantify tissue cytoarchitecture, which has so far been accessible only through histology. Being able to characterize a tissue in vivo would help us define the cytoarchitectonic boundaries and link anatomical and functional information in the cerebral cortex. Our projects include the relationship between diffusion imaging-based brain tissue microstructure, connectivity, and cognition