FlexMol is an Associate Inria Team shared between Nano-D at Inria Grenoble and ChaconLab team at Rocasolano Institute of Physical Chemistry (IQFR-CSIC), Madrid, Spain. Team members include:
- Nano-D:
- Sergei Grudinin, permanent contract, CR CNRS
- https://team.inria.fr/nano-d/team-members/sergei-grudinin/
- expertise in statistical physics and machine learning in application to structural biology
- Karina Machado, visiting professor, FURG Brazil
- http://karina.c3. furg.br
- expertise in bioinformatics and computational biology. Her project is the development of novel computational techniques for flexible protein-ligand docking
- Didier Devaurs, postdoc, Grenoble Alpes University, 12.2018 – 12.2020
- https://csweb. rice.edu/didier-devaurs
- expertise in application of robotics algorithms to the problems of structural biology. His main research project here is linked with the developments for scattering algorithms for flexible molecules
- Isaure Chauvot de Beauchene, CR CNRS since 2016
- https://members.loria.fr/IsaureCB/
- is a specialist in discrete combinatorial methods for RNA flexibility and fragment library-based methods
- Antoine Moniot (French Ministerial bursary), PhD student at Capsid, 10/2018 – 09/2021
- https://team.inria.fr/nano-d/team-members/emilie-neveu/
- will modell 3D protein-RNA complexes by combinatorial fragment assembly
- Maria Kadukova, PhD student, Grenoble Alpes University & MIPT Moscow, 2016-2020
- Maria’s project consists in developing machine-learning algorithms for protein-ligand inter- actions. She is working on the challenge of modeling flexibility of the binding pockets of receptor proteins
- Guillaume Pages, PhD student, Inria, 2016-2019
- Guillaume develops deep-learning algo- rithms for three-dimensional structural molecular data
- Maria Elisa Ruiz (Inria Inria CORDI-s), PhD student at Capsid, 2016 – 11.2019.
- was developing real-space methods for protein-protein docking
- Sergei Grudinin, permanent contract, CR CNRS
- ChaconLab (http://chaconlab.org/):
- Pablo Chacon (Senior Researcher and group leader at IQFR-CSIC, Madrid, Spain)
- is an expert on fast algorithms for macromolecular docking, cryo-EM structure fitting, NMA, and multi-scale simulations
- Jose-Ramon Lopez-Blanco, senior postdoc at IQFR-CSIC (2010-2020), Madrid, Spain
- has extensive experience in structural bioinformatics and in electron microscopy. He received his PhD at UCM studying NMA and its application to macromolecular fitting, where he developed computational tools currently used in the community. His work was also complemented with successful collaborative works with experimental labs including recent Nature and Science articles
- Pablo Solar Rodrıguez, PhD student in Computer engineering, 2017 – 2020, Madrid, Spain.
- His project centers around integrative structural biology
- Pablo Chacon (Senior Researcher and group leader at IQFR-CSIC, Madrid, Spain)
Record of activities :
- Pablo Chacon, Sergei Grudinin, Isaure Chauvot de Beauchene and Maria Elisa Ruiz met during the MASIM 2018 meeting in Paris
- Pablo Chacon, Sergei Grudinin, and Maria Elisa Ruiz met at the CAPRI 2019 conference in Cambridge
- Pablo Chacon visited Nano-D in May 2019 (3 days)
- Pablo Chacon visited Nano-D in September 2019 (3 days)
- Sergei Grudinin and Pablo Chacon visited Capsid team in May 2019 (3 days)
- Maria Kadukova IQFR Madrid in September 2019 (10 days)
- Karina Machado visited IQFR Madrid in September 2019 (10 days)
- Maria Elisa Ruiz visited IQFR Madrid in November 2019 (2 weeks)
Scientific progress :
The associate team has developed 2 novel algorithms that are going to be submitted for publication in the very near future.
- One is an innovative way to describe a multi-level protein flexibil
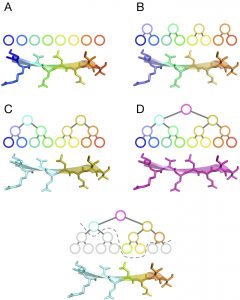 ity using a binary tree.
ity using a binary tree. - Another is a new combination of knowledge-based potentials developed by Nano-D and IQFR applied to protein-ligand interactions at a coarse representation of the protein molecule.
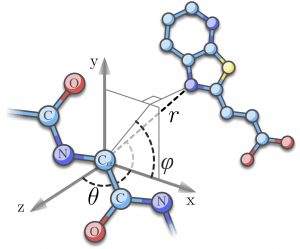
- The associate team has also written an ANR project application.
Production :
None so far.




