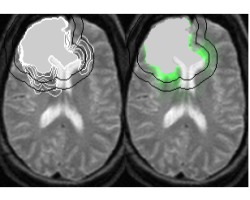
Extrapolating Tumor Invasion Margins for Radiotherapy
In radiotherapy treatment, the constant margin taken around the visible tumor is a very coarse approximation of the invasion margin of cancerous cells. This work tries to solve the problem of adapting the radiotherapy regions to the tumor growth dynamics. The method proposes approximate invasion margins of the tumor based on its growth dynamics. Determining radiotherapy regions based on these invasion margins would increase the effectiveness of the treatment. The low density tumor parts, undetectable by the current imaging techniques, are extrapolated in a magnetic resonance image (MRI). The extrapolation takes into account the underlying tissue structure (grey matter, white matter, fiber directions), the tumor growth dynamics approximated by the Fisher-Kolmogorov model and the segmented tumor in the image.
Contact: Hervé Delingette
Fast and Simple Tensor Processing
 Symmetric positive-definite matrices (or SPD matrices) of real numbers, also called here `tensors’ by abuse of language, appear in many contexts. In medical imaging, their use has become common during the last ten years with the growing interest in Diffusion Tensor Magnetic Resonance Imaging (DT-MRI or simply DTI). SPD matrices also provide a powerful framework to model the anatomical variability of the brain. More generally, they are widely used in image analysis, especially for segmentation, grouping, motion analysis and texture segmentation. They are also used intensively in mechanics, for example with strain or stress tensors. Last but not least, SPD matrices are becoming a common tool in numerical analysis to generate adapted meshes to reduce the computational cost of solving partial differential equations (PDEs) in 3D
Symmetric positive-definite matrices (or SPD matrices) of real numbers, also called here `tensors’ by abuse of language, appear in many contexts. In medical imaging, their use has become common during the last ten years with the growing interest in Diffusion Tensor Magnetic Resonance Imaging (DT-MRI or simply DTI). SPD matrices also provide a powerful framework to model the anatomical variability of the brain. More generally, they are widely used in image analysis, especially for segmentation, grouping, motion analysis and texture segmentation. They are also used intensively in mechanics, for example with strain or stress tensors. Last but not least, SPD matrices are becoming a common tool in numerical analysis to generate adapted meshes to reduce the computational cost of solving partial differential equations (PDEs) in 3D
As a consequence, there has been a growing need to carry out computations with these objects, for instance to interpolate, restore, enhance images of tensors. To this end, one needs to define a complete operational framework. This is necessary to fully generalize to the SPD case the usual statistical tools or PDEs on vector-valued images. In this work, we have proposed a novel and general processing framework for tensors, called ‘Log-Euclidean’. It is based on Log-Euclidean Riemannian metrics, which have excellent theoretical properties, very close to those of the recently-introduced affine-invariant metrics and yield similar results in practice, but with much simpler and faster computations.
This innovative approach is based on a novel vector space structure for tensors. In this framework, Riemannian computations can be converted into Euclidean ones once tensors have been transformed into their matrix logarithms, which makes classical Euclidean processing algorithm particularly simple to recycle.
Contacts: Xavier Pennec.
Related publications can be found here.
Statistical Shape Models Based on Correspondence Probabilities and the EM-ICP Algorithm
The goal of this project is the development of model-based methods for automatic 3D segmentation and recognition of anatomical structures in medical images. A statistical shape model is generated using point cloud representation of the instances. In order to avoid typical problems of direct point-to-point correspondences, a correspondence probability approach is realized when registering the instances (see figures 1 and 2).
Contacts: Xavier Pennec
Related publications can be found here.
A Statistical Study of the Cardiac Diffusion Tensor Images
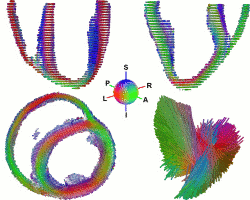 While the main geometrical arrangement of myofibers has been known for decades, its variability between subjects and species still remains largely unknown. Understanding this variability is not only important for a better description of physiological principles but also for the planning of patient-specific cardiac therapies. Furthermore, the knowledge of the relation between the myocardium shape and its myofiber structure is an important and required stage towards the construction of computational models of the heart since the fiber orientation plays a key role when simulating the electrical and mechanical function of the heart. The knowledge about fibre orientation has been recently eased with the use of Diffusion Tensor Imaging (DTI) since there is a correlation between the myocardium fibre structure and diffusion tensors. DTI also has the advantage to provide directly this information in 3D with a high resolution, unfortunately it is not available in vivo due to the cardiac motion. A statistical study of ex vivo cardiac DTI will help in understanding the cardiac fibre structure and in modeling the electromechanical behavior of the heart.
While the main geometrical arrangement of myofibers has been known for decades, its variability between subjects and species still remains largely unknown. Understanding this variability is not only important for a better description of physiological principles but also for the planning of patient-specific cardiac therapies. Furthermore, the knowledge of the relation between the myocardium shape and its myofiber structure is an important and required stage towards the construction of computational models of the heart since the fiber orientation plays a key role when simulating the electrical and mechanical function of the heart. The knowledge about fibre orientation has been recently eased with the use of Diffusion Tensor Imaging (DTI) since there is a correlation between the myocardium fibre structure and diffusion tensors. DTI also has the advantage to provide directly this information in 3D with a high resolution, unfortunately it is not available in vivo due to the cardiac motion. A statistical study of ex vivo cardiac DTI will help in understanding the cardiac fibre structure and in modeling the electromechanical behavior of the heart.
Tensor computing
 The emergence of diffusion tensor imaging (DTI) in the medical imaging community led to challenges in mathematics in order to manipulate 2nd order symmetric positive-defined matrices, so called tensors. In DT-MRI, each voxel of the brain contains a tensor, which is the 2nd order approximation of the brownian motion of water at this specific location. As water tends to move along oriented tissues (such as white matter neural fibers) diffusion tensors are likely to be aligned with the underlying structures. Due to the noise corrupting the data, the tensor field needs adequate post-processing before any further analysis. However, one cannot manipulate tensors like scalars (the tensor space is not a vector space). We used results in differential geometry to manipulate tensors while ensuring to remain on the tensor space, i.e. to keep the positive-defined constrain verified at all time. Applications are: tensor field regularization (PDE, etc.) and shape statistics (see the collaboration Epidaure-LONI).
The emergence of diffusion tensor imaging (DTI) in the medical imaging community led to challenges in mathematics in order to manipulate 2nd order symmetric positive-defined matrices, so called tensors. In DT-MRI, each voxel of the brain contains a tensor, which is the 2nd order approximation of the brownian motion of water at this specific location. As water tends to move along oriented tissues (such as white matter neural fibers) diffusion tensors are likely to be aligned with the underlying structures. Due to the noise corrupting the data, the tensor field needs adequate post-processing before any further analysis. However, one cannot manipulate tensors like scalars (the tensor space is not a vector space). We used results in differential geometry to manipulate tensors while ensuring to remain on the tensor space, i.e. to keep the positive-defined constrain verified at all time. Applications are: tensor field regularization (PDE, etc.) and shape statistics (see the collaboration Epidaure-LONI).
Contacts: Xavier Pennec.
Related publications can be found here.
Statistical evaluation and modeling of human brain variability
 This study is part of the partnership between the Epidaure team and the LONI (Laboratory of Neuro Imaging) at Los Angeles. While our team has been focusing on developing tools adapted to the analysis and registration of multidimensional and multimodal medical images, the LONI has been leading studies to build cerebral atlases of specific diseases (e.g. Alzheimer, schizophrenia) and thus has been collecting anatomical images for years. Today, this amount of images constitutes a unique databasis of more than 500 T1 MRI. Moreover, each images comes with an accurate delineation of 32 pairs of sulci, each sulcal line being segmented by experts who followed a meticulous protocol.
This study is part of the partnership between the Epidaure team and the LONI (Laboratory of Neuro Imaging) at Los Angeles. While our team has been focusing on developing tools adapted to the analysis and registration of multidimensional and multimodal medical images, the LONI has been leading studies to build cerebral atlases of specific diseases (e.g. Alzheimer, schizophrenia) and thus has been collecting anatomical images for years. Today, this amount of images constitutes a unique databasis of more than 500 T1 MRI. Moreover, each images comes with an accurate delineation of 32 pairs of sulci, each sulcal line being segmented by experts who followed a meticulous protocol.
Basically, this work aims at learning the global brain variability from the variability of sulcal lines. The size of the databasis being statistically significant, this work could lead to new results in neuro-anatomy. In the one hand, a better knowledge of the brain variability among a population could help to early detect neurological pathologies and in the other hand, providing a map of the variability could help to guide non-rigid registration algorithms. Indeed, if we are able to predict in which direction each position of the brain statiscally moves, we will be able to better constrain registration algorithms. This work makes an intensive use of a large panel of scientific domains: computer vision, image analysis, statistics and differential geometry to quote just a few.
See the collaboration Asclepios-LONI for further information.
Contacts: Xavier Pennec.
Related publications can be found here.
Study of Meristem Growth
 The meristem is the generative part of the plant. It is a little set of cells located at the tip of each axis. In order to study the plant growth, we need a better description of the meristem. In the context of the ATP (Action Thematique Programmee) Meristeme, a collaraborative project funded by the CIRAD, the goal of this project is to improve the understanding of the meristem growth. We study its cellular organization (the meristem anatomy) and try to understand how this little set of cells can produce the whole arborescence of the tree (the meristem dynamic behavior)
The meristem is the generative part of the plant. It is a little set of cells located at the tip of each axis. In order to study the plant growth, we need a better description of the meristem. In the context of the ATP (Action Thematique Programmee) Meristeme, a collaraborative project funded by the CIRAD, the goal of this project is to improve the understanding of the meristem growth. We study its cellular organization (the meristem anatomy) and try to understand how this little set of cells can produce the whole arborescence of the tree (the meristem dynamic behavior)
This study require the detailed observation of the tissue structure with cellular resolution. Then it is important to develop methods that enable us to observe the inner parts of the organs, in order to analyse and simulate their behavior. To obtain information from the inner cell tissues, we acquire 3D microscopic images in the CIRAD-PHIV in Montpellier, using a multi-photon laser scanning microscope. Then we process these images in order to get 3D segmentations (for static anatomy understanding) and 3D geometrical models of the meristem (in order to compute simulations of the meristem growth)
The next goal consists in following the meristem during its growth, using time-lapse acquisitions, and to get dynamic information from it.
Thanks to Action Thematique Programmee Meristeme (CIRAD / Region Languedoc-Roussillon / IRD) for funding.
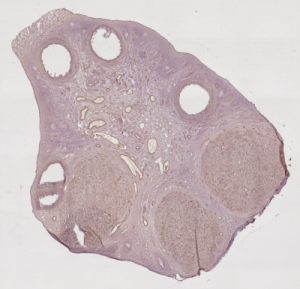
Imaging study of the ovarian function
This work takes place in the Cooperative Research Initiative named REGulation of Ovulation (REGLO). This CRI aims to study the follicular development in mammals, and proposes mathematical models that allow to follow the granulosa cell population, and then to predict the outcome of the follicular development (ovulation or degeneration) with respect to the hormonal environment. To provide the developed models with quantitative data, this work aims at reconstructing a 3-D image of the ewe’s ovary from a series of 2-D stained histologic images.
Contact: Grégoire Malandain
Robust Mosaicing for In Vivo and In Situ Fibered Confocal Microscopy

Fibered confocal microscopy (FCM) is a potential tool for in vivo and in situ optical biopsy. FCM is based on the principle of confocal microscopy which is the ability to reject light from out-of-focus planes and provide a clear in-focus image of a thin section within the sample. This optical sectioning property is what makes the confocal microscope ideal for imaging thick biological samples
The goal of this work is to enhance the possibilities offered by FCM by using image sequence mosaicing techniques in order to widen the field of view (FOV). The image is a mosaic of a sequence acquired ex vivo on a human colon (1500 frames).
Related publications can be found here.
Cardiovascular system segmentation from MRI
In the context of the INRIA national action CARDIOSENSE 3D, this project is about the study of heart segmentation from MRI, using active models and statistical classification. The goal is to achieve the effective segmentation of the atria and proximal arteries in order to complete the current anatomical model where only ventricles are available. The simulation of the electromechanical activity of the heart on this model will participate in diagnosis and therapy of cardiovascular pathologies.
Contacts: Maxime Sermesant
Related publications can be found here
Segmentation of multimodal magnetic resonance images and application to multiple sclerosis
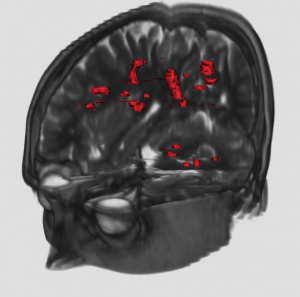 Multiple Sclerosis (MS) is an inflammatory disease of the Central Nervous System (CNS). Predominantly, it is a disease of the “white matter” tissue. In people affected by MS, patches of damage called plaques or lesions appear in seemingly random areas of the CNS white matter. At the site of a lesion, a nerve insulating material, called myelin, is lost. The quantification of the number of lesions, as well as their volume, could be of interest to assess the severity of the disease, and the success of a therapy. These lesions are of different types, and can not be all distinguished in one MR image: therefore, to better characterize them, several sequences are acquired (T1 weighted, with and without Gd, T2-weighted, PD-weighted). We are interested in developing classification and segmentation algorithms that will take advantage of all the available sequences to yield precise quantifications of the lesions in MS patients.
Multiple Sclerosis (MS) is an inflammatory disease of the Central Nervous System (CNS). Predominantly, it is a disease of the “white matter” tissue. In people affected by MS, patches of damage called plaques or lesions appear in seemingly random areas of the CNS white matter. At the site of a lesion, a nerve insulating material, called myelin, is lost. The quantification of the number of lesions, as well as their volume, could be of interest to assess the severity of the disease, and the success of a therapy. These lesions are of different types, and can not be all distinguished in one MR image: therefore, to better characterize them, several sequences are acquired (T1 weighted, with and without Gd, T2-weighted, PD-weighted). We are interested in developing classification and segmentation algorithms that will take advantage of all the available sequences to yield precise quantifications of the lesions in MS patients.
Contact: Grégoire Malandain
Related publications can be found here.
Segmentation of anatomical structures of the lower abdomen with deformable models
The delineation of anatomical structures based on images of the lower abdomen in the frame of dose calculation for conformational radiotherapy is very complex to automatize. We are working on a semi-automatic delineation of these structures in tomodensitometric (CT) images. The method is based on deformable templates whose deformation is guided by the image and by the user as well, in case the latter desires to correct the automatic delineation. In order to validate our approach, we use a set of CT images that have been segmented by medical experts. These hand-made contours act in fact as ”ground truth”, allowing for an objective evaluation of the performance of our algorithm
Contacts: Hervé Delingette
Related publications can be found here
Atlas-based Brain Critical Structures Segmentation for Radiotherapy Planning
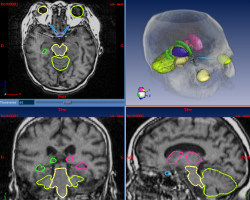
Brain radiotherapy must achieve two goals: the complete irradiation of the tumor, and the preservation of critical structures (brainstem, eyes, optical tracts, etc.). By customizing the shape of the irradiation beam and modulating the irradiation intensity, conformal radiotherapy allows to optimize the irradiation of the tumor and the critical structures. The planning of conformal radiotherapy requires accurate localizations of the tumor and the critical structures.
This work aims to delineate automatically the critical structures of the brain. In order to do this in a specific patient’s image, we use an anatomical atlas containing labels of the structures of the brain. This atlas is registered on the patient image by a global followed by a non rigid registration. The aim of this work is basically to develop a non rigid registration algorithm adapted to anatomical structures registration. The second goal is then to validate the method we propose using quantitative measures on manually delineated patients.
Contacts: Grégoire Malandain.
More details are here.
Quantitative parameters extraction for vascular networks in 3D images
 The purpose of this study is to analyse vascular networks which are recorded using 3D confocal microscopy or other suitable 3D imaging techniques. This includes analysis of the network’s topology, morphometry of individual vessels, and statistical analysis of morphometric properties on the network. To perform such analysis, we must extract a suitable discrete representation of the vascular network from the image data.
The purpose of this study is to analyse vascular networks which are recorded using 3D confocal microscopy or other suitable 3D imaging techniques. This includes analysis of the network’s topology, morphometry of individual vessels, and statistical analysis of morphometric properties on the network. To perform such analysis, we must extract a suitable discrete representation of the vascular network from the image data.
This research takes place in the Microvisu3D Project.
Contacts: Grégoire Malandain
Brain shift modeling
It is usually the case in neurosurgery that pre-operative planning is based on the assumption that anatomical structures do not move between the image acquisition time and the operation time. In reality, the position of brain tissues changes during the operation and significantly decreases the accuracy of the planning made on the pre-operative image.
We propose a bio-mechanical approach based on a finite element model (FEM) of the brain to model this deformation. It takes into account the patient specificity and anatomical cerebral structures to predict the brain deformation.
This study is based on the analysis of 7 cases of Parkinson operations, realized at La Pitie Salpetriere hospital (Paris). It focuses on modeling the static effects of gravity on the brain deformation after dura-mater opening, visible on the post-operative MRI.
More details can be found here
Contacts:Olivier Clatz
Performance evaluation and validation of registration algorithms

The performances of an automatic registration algorithm can be characterized by the accuracy of the result (when the algorithm has converged to the right solution), and the probability of convergence to this correct solution (robustness). Most often, the registration algorithm needs an initial transformation and one prefers to determine a convergence basin in the space of initial transformations where the probability of convergence to the justify solution is close to 100%. Within the accuracy, one can further distinguish between repeatability (or precision) (the error due to the presence of multiple local minima in the immediate vicinity of the optimal solution) and the external error due to the noise on the data.
The purpose of this research theme is to construct the tools to evaluate the performances of registration algorithms on a database of images representative of a clinical application. One of the special aspects of this work is the development of methods to evaluate a lower bound on the registration accuracy without any gold standard, using consistency studies via registration loops.
Related publications are available
Contact: Xavier Pennec
Soft Tissue Modeling for Surgery Simulation
 In collaboration with IRCAD (Institut de Recherche Contre le Cancer de l’Appareil Digestif) located in Strasbourg, we have developed over the past seven years different software technology for the simulation of soft tissue deformation for the simulation of surgery. Surgical simulation aims at training medical resident or surgeons for the practice of video-surgery by providing a software and hardware platform where essential surgical gestures such as suturing, cutting and resection can be simulated in real-time.
In collaboration with IRCAD (Institut de Recherche Contre le Cancer de l’Appareil Digestif) located in Strasbourg, we have developed over the past seven years different software technology for the simulation of soft tissue deformation for the simulation of surgery. Surgical simulation aims at training medical resident or surgeons for the practice of video-surgery by providing a software and hardware platform where essential surgical gestures such as suturing, cutting and resection can be simulated in real-time.
We have developed a simulator for the resection of the liver that includes soft tissue deformation and force-feedback. Among the different scientific difficulties raised by this project, we have focused on the real-time computation of linear and non-linear elastic soft tissue (with transversal anisotropy) but also on the geometric and mechanical aspects of tissue cutting simulation.
Contacts: Hervé Delingette
SPECT imaging in Alzheimer’s Disease

Alzheimer’s disease (AD) is a neuro-degenerative disease that produces among others memory loss, behavioral changes and cognitive impairment. It affects mainly elderly. Because of the aging populations the number of patients will probably rise in the coming years. Therefor powerful diagnostical tools are increasingly important for this disease. Single photon emission computed tomography (SPECT) is being largely used for the study of cerebral blood flow (CBF). These studies provide unique information for the identification of functional abnormalities relevant to Alzheimer’s disease. In collaboration with Professor J. Darcourt of the University of Nice, we are working on the development of diagnosis assisting tools for this type of images. Most of our tools work by comparing images to images of which we already know the diagnosis, and are statistically based.
Reconstruction of a 3-D volume from a series of 2-D images and fusion with a 3-D imaging modality
Within the EC funded project MAPAWAMO we have developed a set of tools that allows us to first reconstruct a 3-D volume (consistent in geometry and in intensity) from a series of 2-D sections (e.g. autoradiographs, histological sections, …). The geometry of such a 3-D may not exactly match the true anatomy of the imaged material. To address this issue, we have also develop a specific methodology that allows to fuse this reconstruction with a 3-D image (e.g. a pre-mortem MR image).
Some related publications can be retrieved through our bibliography pages.
Contact: Grégoire Malandain
Accuracy and robustness of point-surface rigid registration techniques – Application to computer guided oral implantology
 This work is part of an industrial computer-guided surgery project dedicated to oral implantology surgery. Briefly, The operation is planned on a pre-operative CT-Scan and the purpose of such a system is to help the dentist to drill the implant in the predefined position and orientation.
This work is part of an industrial computer-guided surgery project dedicated to oral implantology surgery. Briefly, The operation is planned on a pre-operative CT-Scan and the purpose of such a system is to help the dentist to drill the implant in the predefined position and orientation.
The DentalNavigator system, developed by AREALL, is a per-operative system based on registration. In the CT-Scan image, the teeth and jaw bone surfaces are easily segmented resulting in about 100000 triangulated points. Points on the same structures are measured on the patient resulting in about 10000 unstructured points. After the registration of the 2 surfaces, the system can visually guides the surgeon to the planned position and orientation for drilling.
We investigated several points-surfaces registration techniques and developped tools to compare their speed, robustness and accuracy. We formalised the well-known ICP algorithm and its numerous variants in a statistical framework, and finaly focused on an EM variant and proved it has a multi-scale behaviour. Using a coarse-to-fine approach we resolved robustness problems while improving its accuracy. Using a simple decimation technique, we down-sampled our sets of points and increased the algorithm speed while preserving its properties. Current work is going on the theoretical prediction of algorithm accuracy and robustness.
Another part of this work is about the probabilistic modeling of noisy curves and surfaces. We are currently trying to explain the tensor-voting techniques developped by Medioni and his team, and hope to develop a statistical framework useful for surface reconstruction, fusion and registration.
A posteriori validation of pre-operative planning in functional neurosurgery by quantification of brain pneumocephalus
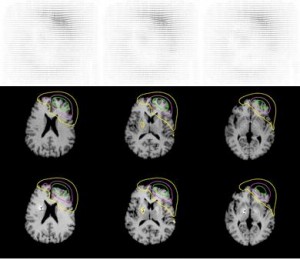
In MR image guided brain surgery, the techniques are often based on the use of volumetric pre-operative MR acquisitions. This implicitely assumes that a pre-operative acquisition gives a faithful and precise representation of the brain anatomy during the intervention. Thus a major limit of these techniques is the development of brain deformation during the surgical intervention, leading to anatomical differences, which can be significative, with the pre-operative MR images.
One example of this type of procedures is functional neurosurgery for Parkinson’s disease. This intervention is based on the stereotactic introduction of electrodes in a small, deeply located, nucleus of the brain, called the subthalamic nucleus, which is targeted on pre-operative stereotactic MR acquisitions. During the intervention, which is performed outside the MR unit, an electrophysiological and clinical study is performed with the electrodes to check the pre-operatively determined target position. This exploration is time-consuming and can lead to the development of a pneumocephalus (presence of air in the intracanial cavity) because of CSF leak. This induces a brain shift which can yield a significative deformation of the entire brain and thus cause potential errors in the pre-operatively determined position of the stereotactic targets.
We have developed in collaboration with the Neuroradiology Dept. of La Salpetriere hospital (Paris) a method to quantify brain deformation from pre and immediate post-operative MR acquisitions, based on rigid and non rigid registrations. The novel non rigid registration algorithm used in this study is based on a true free form deformation modelisation coupled with a mixed regularization. This method will allow to quantify the deformation occurred around the stereotactic targets and lead to a posteriori validation of the pre-operative planning.
Some publications are available.
Motion Estimation and Analysis in Echocardiographic data
 The purpose of this work is the estimation and analysis of displacement fields of the myocardium. The motion will be estimated in 2D and 3D echocardiographic images with variational techniques. The analysis is performed with level sets methods. First estimations were obtained with 2D echocardiographic images using Doppler Tissue Imaging. Then motion analysis will be improved by using a priori information such as a biomechanical model of the heart.
The purpose of this work is the estimation and analysis of displacement fields of the myocardium. The motion will be estimated in 2D and 3D echocardiographic images with variational techniques. The analysis is performed with level sets methods. First estimations were obtained with 2D echocardiographic images using Doppler Tissue Imaging. Then motion analysis will be improved by using a priori information such as a biomechanical model of the heart.
Anatomo-Functional Parcellation of the Brain

In this study, we develop a method for brain parcellation.
This work is part of collaboration between the Epidaure Research Project and the CEA-SHFJ at Orsay.
Some publications are available.
Registration of post mortem macroscopic optical brain slice images with MR scans of the same patient
 In this study, we develop a method for registration of macroscopic optical images with MR images of the same patient. This forms a key part of a series of procedures to allow post mortem findings to be accurately registered with MR images, and more generally provides a method for 3D mapping of the distribution of pathological changes throughout the brain. The first stage of the method involves a 3D reconstruction of 2D brain slices. The second stage consists in the registration of the reconstructed volume with corresponding MR images. This work is part of the QAMRIC european project.
In this study, we develop a method for registration of macroscopic optical images with MR images of the same patient. This forms a key part of a series of procedures to allow post mortem findings to be accurately registered with MR images, and more generally provides a method for 3D mapping of the distribution of pathological changes throughout the brain. The first stage of the method involves a 3D reconstruction of 2D brain slices. The second stage consists in the registration of the reconstructed volume with corresponding MR images. This work is part of the QAMRIC european project.
Some publications are available.
Automated Segmentation of Anatomical Structures in Brain MRI
 Effective identification and labeling of anatomical structures in Magnetic Resonance Imaging (MRI) proves to be challenging, given the wide variety of shapes and intensities each structure can present. Automated image segmentation can be used to assist medical analysis, and calls for high precision as the quality of the diagnosis often depends on how accurately those structures can be identified. Similarly, brain atlases, which can provide a precise quantitative framework for multimodality brain mapping, turn out to be rather tedious to build, as many components typically have to be interactively outlined. Thus, automated segmentation system are powerful tools to help in drawing consistent diagnosis from a number of images, to classify pictorial data, or collect statistical information on anatomical variability.
Effective identification and labeling of anatomical structures in Magnetic Resonance Imaging (MRI) proves to be challenging, given the wide variety of shapes and intensities each structure can present. Automated image segmentation can be used to assist medical analysis, and calls for high precision as the quality of the diagnosis often depends on how accurately those structures can be identified. Similarly, brain atlases, which can provide a precise quantitative framework for multimodality brain mapping, turn out to be rather tedious to build, as many components typically have to be interactively outlined. Thus, automated segmentation system are powerful tools to help in drawing consistent diagnosis from a number of images, to classify pictorial data, or collect statistical information on anatomical variability.
In collaboration with the Laboratory of NeuroImaging, at the UCLA School of Medicine in Los Angeles, within the framework of an Associated Team, we propose to develop a fully automated segmentation system to extract deep and surface anatomical structures from brain MRI. We approach the issue of boundary finding as a process of fitting a series of statistically controlled deformable templates to the target contour.
Our goal is to determine and analyze the features of template matching algorithms that play a determining role in terms of their efficiency and accuracy.
Rigid and non-rigid registration for motion compensation and subject comparison in fMRI studies
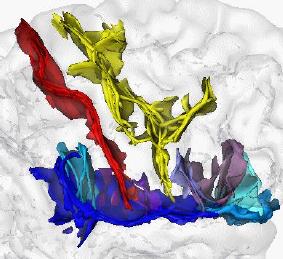
This research theme is implemented within the framework of an INRIA “development action”, a concerted action between different INRIA and other French teams aimed at producing new software. The goal is to adapt and evaluate the different non rigid registration algorithms already developed at INRIA, mainly at EPIDAURE, ODYSSEE and VISTA, in order to fulfill the needs of fMRI investigators, in our case the CEA/SHFJ/DRM and INSERM U494 (IMQ) teams.
Three main problems are investigated:
- Motion compensation during an fMRI acquisition sequence. It is important to compensate very accurately (or at least detect) for the subject motion between images as a very small registration error may lead to spurious activation detections.
- Inter-subject registration to fuse functional information from different subjects. There is currently no satisfying method to establish point to point correspondences between two brains. The main idea developed here is to enrich iconic (i.e. intensity-based) methods with geometric constraints based on anatomical sulcal correspondences.
- The validation of this inter-subject registration. As there is no ground truth, the evaluation of the previous algorithms is very difficult. One interesting idea is to estimate the variability of anatomically stable functional activations across subjects and to show the the inter-subject registration drastically reduce this variability, hopefully removing all the anatomical variability to leave only the functional one.
More information on the action IRMf home page.
Contact: Xavier Pennec
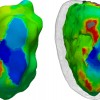
Elaboration of a three-dimensional, functional and registrable atlas of the human basal ganglia
 In order to allow accurate pre-operative localisation of functional targets in functional neurosurgery, we aim at constructing a three dimensional registrable cartography of the basal ganglia, based on histology. For doing this, a {\em post mortem} MR study was conducted on a cadaver’s head, and the brain was then extracted and processed for histology. The post mortem MR image will allow to report the cartography on the patient’s anatomy, by its registration with the patient’s MR image.
In order to allow accurate pre-operative localisation of functional targets in functional neurosurgery, we aim at constructing a three dimensional registrable cartography of the basal ganglia, based on histology. For doing this, a {\em post mortem} MR study was conducted on a cadaver’s head, and the brain was then extracted and processed for histology. The post mortem MR image will allow to report the cartography on the patient’s anatomy, by its registration with the patient’s MR image.
Elaboration of this atlas consists in co-registering the histological and {\em post mortem} MR data of the same subject. First, realignment of the histological sections into a reliable three dimensional volume is performed. Then the reconstructed volume is registered with the post mortem MR image. To insure three dimensional integrity of the histological reconstructed volume, a reference volume is first constructed from photographs of the unstained surface of the frozen brain. This reference is then used as an intermediate volume for, on the one hand, independant alignment of each histological section with its corresponding optical section and on the other hand, three dimensional registration with the post mortem MR image.
This work is part of a collaboration between the U.289 group at INSERM (Paris) and the Neuroradiology Dept. of La Salpetriere hospital (Paris). It was partially supported by Medtronic Inc. (Denver, USA).
Some publications are available.
Augmented Reality for Abdominal Surgery
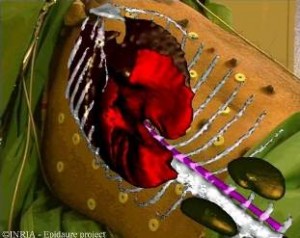
In order to improve surgical accuracy, radiologists and surgeons can use a new imaging tool : Augmented Reality. It deals with superimposing a virtual model built from the pre-operative image on a video image of the patient . In collaboration with IRCAD (Institut de Recherche contre les Cancers de l’Appareil Digestif), we aim at creating an augmented reality system for the destruction of liver tumors with the use of radio-fequency. The purpose is to superimpose on extern video images several tridimensional reconstructions of the liver, the tumors and some other organs. We have developed a new point-based 3D/2D registration method and a visualisation software which lead us to our first result (see image). The 3D models come from segmented CT-scans and the registration is made thanks to radio-opaque fiducials on the patient’s skin. We now have to validate the efficiency and accuracy of this method on porcins and mannequin.
Automatic Detection and Quantification of Evolving Processes in 3D Medical Images
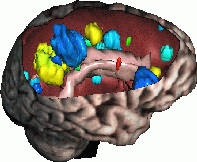 This work has been conducted mainly for the analysis of MR images of patients with Multiple Sclerosis. The goal (de circonstance!) is the design of new tools to help the clinicians to detect and locate new lesions, and also to measure objectively their evolution through time. To achieve this, we develop several approches (cf. publications). This work is done in collaboration with Harvard Medical School (Pr. Ron Kikinis) and with CHU of Nice (Pr. Chatel, Dr. C. Lebrun). Our work on automatic detection and quantification of evolving lesions deals essentially with:
This work has been conducted mainly for the analysis of MR images of patients with Multiple Sclerosis. The goal (de circonstance!) is the design of new tools to help the clinicians to detect and locate new lesions, and also to measure objectively their evolution through time. To achieve this, we develop several approches (cf. publications). This work is done in collaboration with Harvard Medical School (Pr. Ron Kikinis) and with CHU of Nice (Pr. Chatel, Dr. C. Lebrun). Our work on automatic detection and quantification of evolving lesions deals essentially with:
- pre-processings on MRI series (registration, intensity corrections, interpolation …)
- analysis of MRI evolutions. 2 methods have been developped: one based on the computation and on the analysis of a vector field of apparent displacement between 2 MRIs; the other based on a statistical analysis of the intensity evolution of each anatomical point over time.
A demonstration is also available.






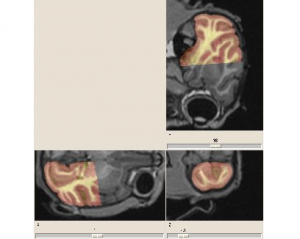
 Non invasive cardiac personalisation
Non invasive cardiac personalisation
 Simulation of ventricular tachycardia re-entry circuit
Simulation of ventricular tachycardia re-entry circuit
 heartMeshFine
heartMeshFine
 Electrophysiology
Electrophysiology
 Cardiac Fibres from in vivo Diffusion Tensor Imaging
Cardiac Fibres from in vivo Diffusion Tensor Imaging
