 Up to date information at my official page.
Up to date information at my official page.
My activity concerns the development and study of computational and statistical methods for the analysis of biomedical data and brain images. I’m also interested in statistical analysis in clinical trials of disease modifying drugs. I’m currently editorial board member of Nature Scientific Report (Neurology Panel), and I worked as statistical consultant for the European clinical project Pharmacog. I was awarded in 2015 with the second ex-aequo prize for the ERCIM Cor Baayen Award.
For an up-to-date list of my publications, please take a look to my Google Scholar page.
NEWS:
05/2018. A new paper accepted to ICML 2018!
Marco Lorenzi & Marizio Filippone. Constraining the Dynamics of Deep Probabilistic Models. https://arxiv.org/abs/1802.05680
A new paper accepted in PNAS!
02/2018. Marco Lorenzi, Andre Altmann, Boris Gutman, Selina Wray, Charles Arber, Derrek P. Hibar, Neda Jahanshad, Jonathan M. Schott, Daniel C. Alexander, Paul M. Thompson and Sebastien Ourselin. Susceptibility of brain atrophy to TRIB3 in Alzheimer’s disease: Evidence from functional prioritization in imaging genetics. Proceedings of the National Academy of Sciences of the United States of America (PNAS). Accepted on Feb 2018.
Selected research topics
- Imaging Genetics
One of my main research axis consists in studying the relationship between brain phenotypes quantified by neuroimaging (such as atrophy, or connectivity) and individual’s genetic profile. This challenging research field requires the development of efficient and powerful statistical models for studying multivariate patterns of associations between imaging and genetics data. The circle plot on the left shows the genetic locations significantly associated to the brain cortical thinning shown on the right hand side.
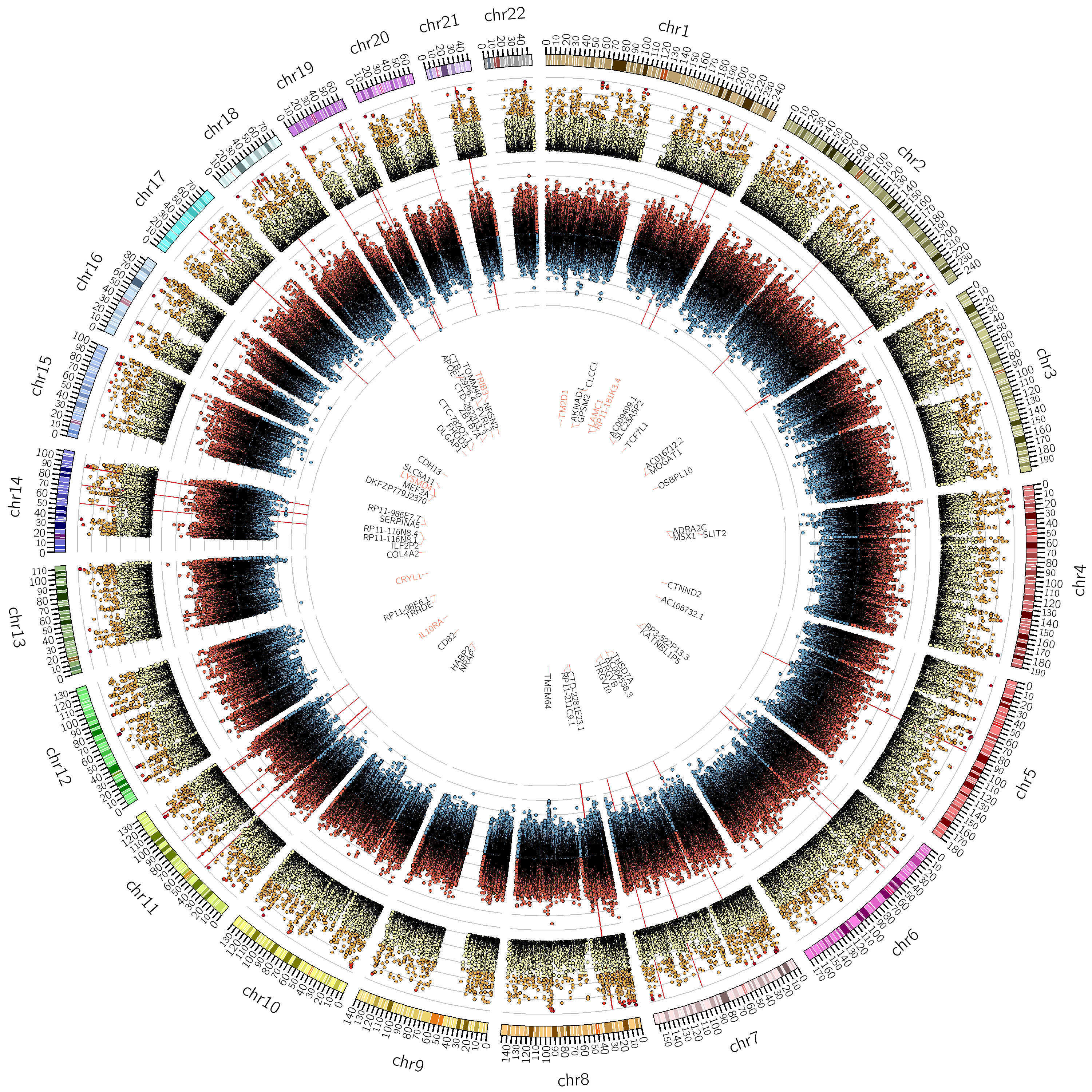 |
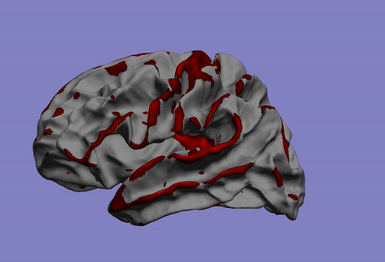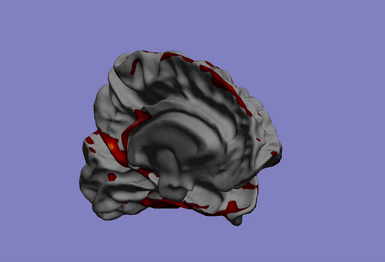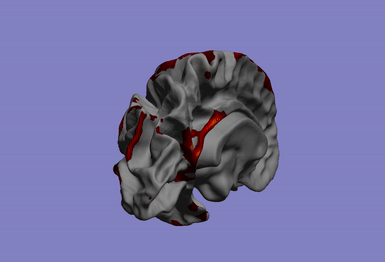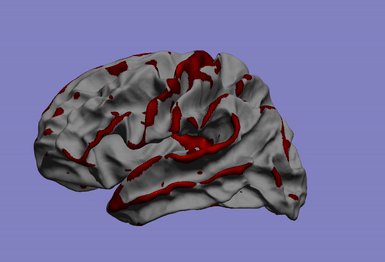 |
02/2018 UPDATE: The work is accepted for publication in PNAS! The experimental material is now available (DOI:10.6084/m9.figshare.5914375.v1).
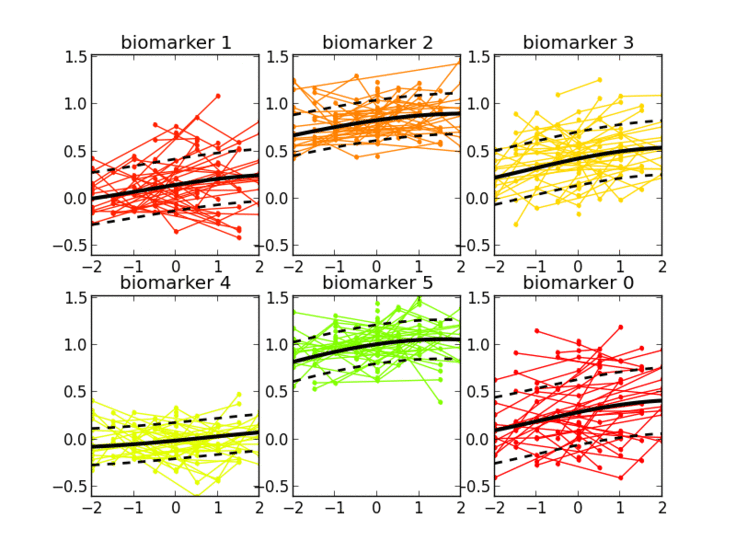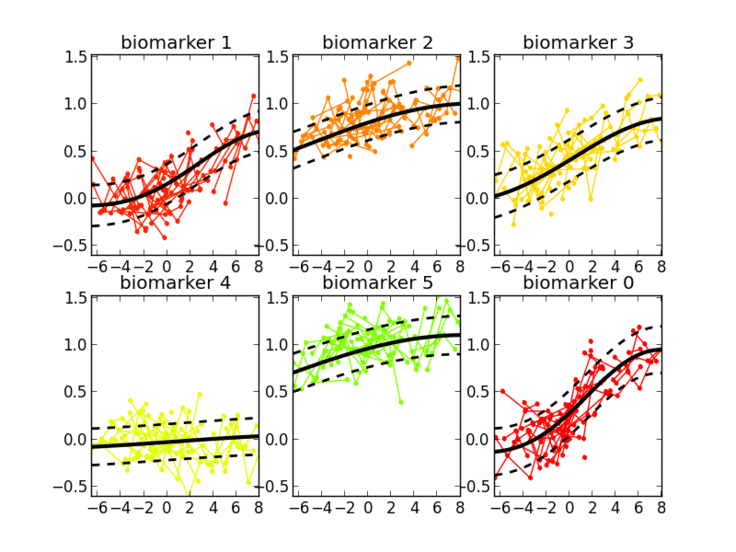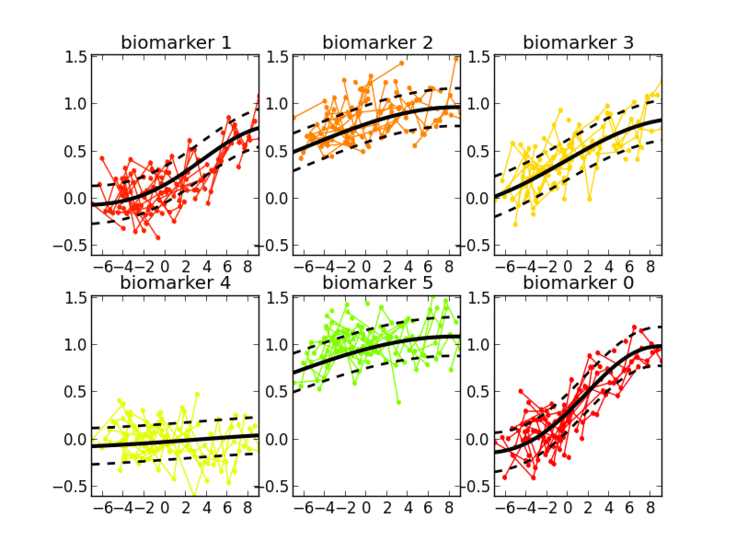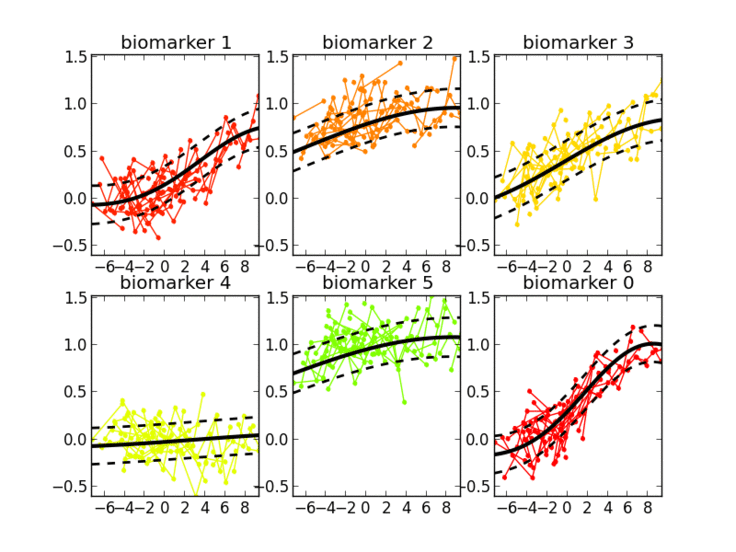
- Gaussian Processes for temporal analysis of clinical data
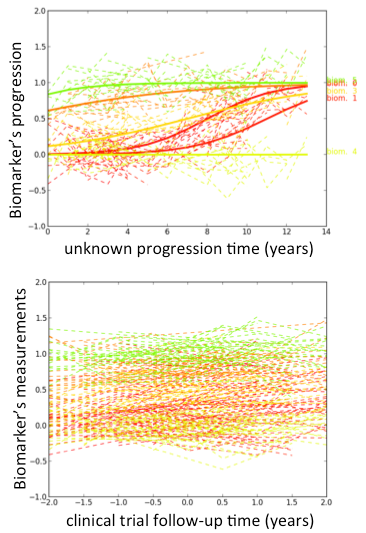
I’m interested in the study of learning algorithms for modeling and predicting longitudinal changes in imaging, clinical and biological data. We can think of time series of patients’ data as measurements from an underlying disease progression trajectory spanning several years (left figure, top). Yet, when we observe the patients during a clinical trial, the disease time information is unknown (left figure bottom). This fascinating research topic aims at reconstructing the natural history of a pathology from individual time series of clinical data (right figure).
12/2017 UPDATE: The NeuroImage paper is finally available. Please contact me if you are interested in testing the software.
- Statistical learning of multimodal image variation
I’ve been working in the analysis of the joint variation between multimodal brain imaging modalities, such as the brain atrophy measured in magnetic resonance images (MRIs), and the hypometabolism quantified by FDG-PET radiotracers. The video shows that the non-local correlation between atrophy and hypomethabolism in Alzheimer’s patients is identified by localised subcortical temporal atrophy and ventricular enlargement, a pattern mostly associated with the hypometabolism of the temporal/parietal cortex.
- Non-linear image registration
I’m interested in the development and study of non-linear registration algorithms to reliably detect morphological changes. The figure below shows the parameters of the diffeomorphic transformation of longitudinal brain atrophy in Alzheimer’s disease (left), and the associated spatio-temporal model of brain changes (right). We note that the registration model accurately describes the global brain atrophy, as well as the ventricular expansion and hippocampal atrophy.
LCC-logDemons is an accurate and robust diffeomorphic image registration framework addressed to the whole medical imaging community. The software is released with free license for research purposes. LCC-logDemons is open source and is based on ITK, a popular C++ medical imaging library.
Bibliography
International journals
[1] Sebastiano Ferraris , Hannes van der Merle, Lennart Van Der Veeken, Herran Prados, Juan Eugenio Iglesias, Andrew Melbourne, Marco Lorenzi, Marc Modat, Willy Gsell, Jan Deprest, Tom Vercauteren. A magnetic resonance multi-atlas for the neonatal rabbit brain. NeuroImage, Accepted on May 2018. code [link]
[2] Marzia A Scelsi, Raiyan R Khan, Marco Lorenzi, Leigh Christopher, Michael D Greicius, Jonathan M Schott, Sebastien Ourselin, Andre Altmann . Genetic study of multimodal imaging Alzheimer’s disease progression score implicates novel loci. Brain, Volume 141, Issue 7, 1 July 2018, Pages 2167–2180 [link]
[3] Marco Lorenzi, Andre Altmann, Boris Gutman, Selina Wray, Charles Arber, Derrek P. Hibar, Neda Jahanshad, Jonathan M. Schott, Daniel C. Alexander, Paul M. Thompson and Sebastien Ourselin. Susceptibility of brain atrophy to TRIB3 in Alzheimer’s disease: Evidence from functional prioritization in imaging genetics. Proceedings of the National Academy of Sciences of the United States of America (PNAS).
[4] Marco Lorenzi, Maurizio Filippone, Giovanni B. Frisoni, Daniel C. Alexander, Sebastien Ourselin. Probabilistic disease progression modeling to characterize diagnostic uncertainty: application to staging and prediction in Alzheimer’s disease. NeuroImage, S1053-8119(17)30706-1, 2017. [link]
[5] Alex F. Mendelson, Maria A. Zuluaga, Marco Lorenzi, Brian F. Hutton, Sebastien Ourselin, (2017). Selection bias in the reported performances of AD classification pipelines. NeuroImage: Clinical, 14, 400-416, 2017. [link]
[6] Bishesh Khanal, Marco Lorenzi, Nicholas Ayache, Xavier Pennec. A biophysical model of brain deformation to simulate and analyze longitudinal MRIs of patients with Alzheimer’s disease. NeuroImage, 134, 35-52, 2017. [link]
[7] Mehdi Hadj-Hamou, Marco Lorenzi, Nicholas Ayache, Xavier Pennec. Longitudinal analysis of image time series with diffeomorphic deformations: a computational framework based on stationary velocity fields. Frontiers in Neuroscience, 10, 236, 2016. [link]
[8] Marco Lorenzi , Ivor J. Simpson, Alex F. Mendelson, Sjoerd B. Vos, M. Jorge Cardoso, Marc Modat, Jonathan Schott, Sebastien Ourselin. Multimodal Image Analysis in Alzheimer’s Disease via Statistical Modelling of Non-local Intensity Correlations. Scientific Reports (NPG), 6, 2261, 2016. [link]
[9] David M. Cash, Chris Frost, Leonardo O. Iheme, Devrim Unay, Melek Kandemir, Jurgen Fripp, Olivier Salvado, Pierrick Bourgeat, Martin Reuter, Bruce Fischl, Marco Lorenzi, Giovanni B. Frisoni, Xavier Pennec, Ronald K. Pierson, et al. Assessing atrophy measurement techniques in dementia: Results from the MIRIAD atrophy challenge. NeuroImage, 123: 149-164, 2015. [link]
[10] Marco Lorenzi , Nicholas Ayache and Xavier Pennec. Regional Flux analysis for discovering and quantifying anatomical changes: An application to the brain morphometry in Alzheimer’s disease. NeuroImage, 115:224-234, 2015. [link]
[11] Marco Lorenzi, Xavier Pennec, Giovanni B. Frisoni and Nicholas Ayache. Disentangling Normal Aging from Alzheimer’s Disease in Structural MR Images. Neurobiology of Aging, 36:S42-S52, 2015. [link]
[12] Antonello Preti, Cristina Muscio, Marina Boccardi, Marco Lorenzi, Giovanni de Girolamo, Giovanni B. Frisoni. Impact of alcohol consumption in healthy adults: A magnetic resonance imaging investigation. Psychiatry Research: Neuroimaging, 224 (2), 96-103, 2014. [link]
[13] Marco Lorenzi and Xavier Pennec. Efficient Parallel Transport of Deformations in Time Series of Images: from Schild’s to Pole Ladder. Journal of Mathematical Imaging and Vision, 50(1-2):5-17, 2014. [link]
[14] Marco Lorenzi, Nicholas Ayache, Giovanni B. Frisoni, and Xavier Pennec. LCC-Demons: a robust and accurate symmetric diffeomorphic registration algorithm. NeuroImage, 81(1):470-483, 2013. [link]
[15] Marco Lorenzi and Xavier Pennec. Geodesics, Parallel Transport & One-parameter Subgroups for Diffeomorphic Image Registration. International Journal of Computer Vision, 105(2):111-127, 2013. [link]
[16] Roberta Rossi, Michela Pievani, Marco Lorenzi, Marina Boccardi, Rossella Beneduce, Stefano Bignotti, Genoveffa Borsci, Maria Cotelli, Panteleimon Giannakopoulos, Laura R. Magni, Luciana Rillosi, Sandra Rosini, Giuseppe Rossi, and Giovanni B. Frisoni. Structural brain features of borderline personality and bipolar disorders. Psychiatry Research: Neuroimaging, 213(2):83-91, 2013. [link]
[17] Marco Lorenzi, Alberto Beltramello, Nicola Mercuri, Elisa Canu, Giada Zoccatelli, Francesca Pizzini, Franco Alessandrini, Maria Cotelli, Sandra Rosini, Daniela Costardi, Carlo Caltagirone, and Giovanni B. Frisoni. Effect of memantine on resting state default mode network activity in Alzheimer’s disease. Drugs and Aging, 28(3):205-217, 2011.
[18] Marco Lorenzi, Michael Donohue, Donata Paternico, Cristina Scarpazza, Susanne Ostrowitzki, Olivier Blin, Elaine Irving, and Giovanni B. Frisoni. Enrichment through biomarkers in clinical trials of Alzheimer’s drugs in patients with mild cognitive impairment. Neurobiology of Aging, 31(8):1443-1451, 2010.
[19] Anna Caroli, Marco Lorenzi, Cristina Geroldi, Flavio Nobili, Barbara Paghera, Mario Bonetti, Maria Cotelli, Giovanni B. Frisoni. Metabolic compensation and depression in Alzheimer’s disease. Dementia and geriatric cognitive disorders, 29(1):37-45, 2010.
[20] Giovanni B. Frisoni, Marco Lorenzi, Anna Caroli, Nina Kemppainen, Kjell Nagren, and Juha O. Rinne. In vivo mapping of amyloid toxicity in Alzheimer’s disease. Neurology, 72(17): 1504-1511, 2009.
Reviewed international conferences with proceedings
[21] Marco Lorenzi, Maurizio Filippone. Constraining the Dynamics of Deep Probabilistic Models. Proceedings of the 35th International Conference on Machine Learning (ICML), PMLR 80:3233-3242, 2018. Note: Oral podium presentation. Acceptance rate: ~25%.
[22] Juan Eugenio Iglesias, Marco Lorenzi, Sebastiano Ferraris, Loic Peter, Marc Modat, Allison Stevens, Bruce Fischl, Tom Vercauteren: “Model-based refinement of nonlinear registrations in 3D histology reconstruction”, MICCAI 2018. Acceptance rate: ~25%. code
[23] Răzvan Valentin Marinescu, Arman Eshaghi, Marco Lorenzi, Alexandra L Young, Neil P Oxtoby, Sara Garbarino, Timothy J Shakespeare, Sebastian J Crutch, Daniel C Alexander. A Vertex Clustering Model for Disease Progression: Application to Cortical Thickness Images. Information Processing in Medical Imaging (IPMI), 10265: 134-145, Springer, LNCS, 2017. Note: Oral podium presentation. Acceptance rate: ~30%
[24] Marco Lorenzi, Boris Gutman, Paul M Thompson, Daniel C Alexander, Sebastien Ourselin, Andre Altmann. Secure multivariate large-scale multi-centric analysis through on-line learning: an imaging genetics case study. 12th International Symposium on Medical Information Processing and Analysis, 1016016, International Society for Optics and Photonics, 2017. Note: Oral podium presentation
[25] Eliza Orasanu, Pierre-Louis Bazin, Andrew Melbourne, Marco Lorenzi, Herve Lombaert, Nicola J Robertson, Giles Kendall, Nikolaus Weiskopf, Neil Marlow, Sebastien Ourselin. Longitudinal analysis of the preterm cortex using multi-modal spectral matching. Medical Image Computing and Computer Aided Intervention (MICCAI), 255-263, Springer, LNCS, 2016. Note: Acceptance rate: ~30%
[26] Sebastiano Ferraris, Marco Lorenzi, Pankaj Daga, Marc Modat, Tom Vercauteren. Accurate small deformation exponential approximant to integrate large velocity fields: Application to image registration. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, 17-24, 2016.
[27] Marco Lorenzi, Boris A. Gutman, Derrek P. Hibar, Andre Altmann, Neda Jahanshad, Paul M. Thompson and Sebastien Ourselin. Partial Least Squares Modelling for Imaging genetics in Alzheimer’s Disease: Plausibility and Generalization. IEEE International Symposium on Biomedical Imaging (ISBI), 2016.
[28] Marco Lorenzi, Gabriel Ziegler, John Ashburner, Daniel Alexander and Sebastien Ourselin. Efficient Gaussian process Based Model of Spatio-Temporal Changes in Time Series of Images. Information Processing in Medical Imaging (IPMI), 24:626-37, Springer, LNCS, 2015. Note: Acceptance rate: ~30%
[29] Marco Lorenzi , Gabriel Ziegler, Daniel C. Alexander, and Sebastien Ourselin. Modelling Non-Stationary and Non-Separable Spatio-Temporal Changes in Neurodegeneration via Gaussian Process Convolution, 1st ICML Workshop on Machine Learning Meets Medical Imaging. 9487, Springer, LNCS, 2015. Note: Oral podium presentation.
[30] Boris A. Gutman, Tom P. Fletcher, Jorge M. Cardoso, Greig M. Fleishman, Marco Lorenzi, Paul M. Thompson, Sebastien Ourselin. A Riemannian Framework for Intrinsic Comparison of Closed Genus-Zero Shapes. Information Processing in Medical Imaging (IPMI), 24:205-18, Springer, LNCS, 2015. Note: Oral podium presentation. Acceptance rate: ~30%
[31] Bishesh Khanal, Marco Lorenzi, Nicholas Ayache and Xavier Pennec. A biophysical model of shape changes due to atrophy in the brain with Alzheimer’s disease . Medical Image Computing and Computer Aided Intervention (MICCAI), 17:41-8, Springer, LNCS, 2015. Note: Acceptance rate: ~30%
[32] Marco Lorenzi, Bjoern Menze, H., Marc Niethammer, Nicholas Ayache, and Xavier Pennec. Sparse Scale-Space Decomposition of Volume Changes in Deformations Fields . Medical Image Computing and Computer Aided Intervention (MICCAI), 328-335, Springer, LNCS, 2013. Note: Acceptance rate: ~30%
[33] Marco Lorenzi and Xavier Pennec. Parallel Transport with Pole Ladder: Application to Deformations of time Series of Images. In GSI2013 – Geometric Science of Information, Springer, LNCS, 8085, 68-75, 2013. Note: Oral podium presentation
[34] Marco Lorenzi, Nicholas Ayache, and Xavier Pennec. Regional flux analysis of longitudinal atrophy in Alzheimer’s disease. In Medical Image Computing and Computer Aided Intervention (MICCAI), 7510:739-746, Springer, LNCS, 2012. Note: Oral podium presentation. Acceptance rate: ~30%.
[35] Marco Lorenzi, Giovanni B. Frisoni, Nicholas Ayache, and Xavier Pennec. Probabilistic Flux Analysis of Cerebral Longitudinal Atrophy. In MICCAI workshop on Novel Imaging Biomarkers for Alzheimer’s Disease and Related Disorders (NIBAD 12), 256-265, 2012. Note: Oral podium presentation.
[36] Marco Lorenzi, Nicholas Ayache, Giovanni B. Frisoni, and Xavier Pennec. Mapping the effects of Aß142 levels on the longitudinal changes in healthy aging: hierarchical modeling based on stationary velocity fields. In Medical Image Computing and Computer Aided Intervention (MICCAI), 6892: 663-670, Springer, LNCS, 2011. Note: Acceptance rate: ~30%.
[37] Marco Lorenzi, Nicholas Ayache, and Xavier Pennec. Schild’s Ladder for the parallel transport of deformations in time series of images. Information Processing in Medical Imaging (IPMI), 6801: 463-474, Springer, LNCS, 2011. Note: Honorable Mention (runner-up) for the Erbsmann Award. Oral podium presentation, acceptance Rate: ~30%
[38] Marco Lorenzi and Xavier Pennec. Geodesics, parallel transport & one-parameter subgroups. In 3rd MICCAI workshop on Mathematical Foundations of Computational Anatomy Workshop, September 2011. Note: Oral podium presentation
[39] Marco Lorenzi, Nicholas Ayache, Giovanni B. Frisoni, and Xavier Pennec. 4D registration of serial brain MRIs images: a robust measure of changes applied to Alzheimer’s disease. In MICCAI Workshop on Spatio- Temporal Image Analysis for Longitudinal and Time-Series Image Data, 2010. Note: Oral Podium Presentation, best presentation award
Book chapters
[40] Marco Lorenzi and Xavier Pennec. Discrete Ladders for Parallel Transport in Transformation Groups with an Affine Connection Structure. Geometric Theory of Information Signals and Communication Technology, 243-271, Springer, 2014.
Oral Podium Presentation in Clinical Conferences
[41] Marco Lorenzi, Helene Barelli. Blood Levels of Omega 3 and 6 across the Progression of Alzheimer’s Disease. In Alzheimer’s Association International Conference, Chicago, US, 2018.
[42] Marco Lorenzi, Maurizio Filippone, Daniel C Alexander, Sebastien Ourselin. Modeling and prediction of the natural history of neurodegeneration from longitudinal trial data. In Alzheimer’s Association International Conference, London, UK, 2017.
[43] Marco Lorenzi, Boris A Gutman, Andre Altmann, Derrek P Hibar, Neda Jahanshad, Daniel C Alexander, Paul M Thompson, Sebastien Ourselin. Linking gene pathways and brain atrophy in Alzheimer’s disease. In Alzheimer’s Association International Conference, Toronto, CA, 2016. Note: Travel Fellowship Award
[44] Marco Lorenzi , Nicholas Ayache, and Xavier Pennec, Giovanni B. Frisoni. Regional flux analysis of longitudinal atrophy in Alzheimer’s disease. In Alzheimer’s Association International Conference, Copenhagen, DK, 2014. Note: Travel Fellowship Award
[45] Marco Lorenzi, Nicholas Ayache, Xavier Pennec, Giovanni B. Frisoni. Disentangling the normal aging from the pathological Alzheimer’s disease progression on cross-sectional structural MR images. International Conference on Clinical Trials on Alzheimer’s Disease (CTAD), Monte Carlo, Monaco, 2012.
[46] Marco Lorenzi, Michael Donohue, Donata Paternic, Cristina Scarpazza, Susan Ostrowitzki, Olivier Blin, Elain Irving, Giovanni B. Frisoni. Enrichment through biomarkers in clinical trials of Alzheimer’s drugs in patients with mild cognitive impairment. International Conference on Clinical Trials on Alzheimer’s Disease (CTAD), Toulouse, France, 2010.
International Clinical Conference Abstracts
[47] Marco Lorenzi, Ivor J. Simpson, Alex F. Mendelson, Sjoerd B. Vos, M. Jorge Cardoso, Marc Modat, Jonathan Schott, Sebastien Ourselin. Multimodal Image Analysis in Alzheimer’s Disease via Statistical Modelling of Non-local Intensity Correlations . In Alzheimer’s Association International Conference, Washington DC, USA, 2015.
[48] Marco Lorenzi, Martina Bocchetta, Nicholas Ayache, Xavier Pennec, and Giovanni B. Frisoni. Conversion to MCI in healthy individuals with abnormal CSF Aß142 levels is associated with specific longitudinal morphological changes. In Alzheimer’s Association International Conference, Boston, USA, 2013. Note: Travel Fellowship Award




 Non invasive cardiac personalisation
Non invasive cardiac personalisation
 Simulation of ventricular tachycardia re-entry circuit
Simulation of ventricular tachycardia re-entry circuit
 heartMeshFine
heartMeshFine
 Electrophysiology
Electrophysiology
 Cardiac Fibres from in vivo Diffusion Tensor Imaging
Cardiac Fibres from in vivo Diffusion Tensor Imaging
