In this project, we design and deploy some computational tools to perform neuroimaging-genetics association studies at a large scale.
Unveiling the relationships between genetic variability and brain structure and function is one of the main challenges in neuroscience, which can be partly addressed through the information conveyed by high-throughput genotyping on the one hand, and neuroimaging data on the other hand. Finding statistical associations between these different variables is important in order to find relevant biomarkers for various brain diseases and improve patient handling. Due to the huge size of the datasets involved and the requirement for tight bounds on statistical significance, such statistical analysis are particularly demanding and cannot be performed easily at a large scale with standard software and computational tools. In ICOGEN, we design and deploy some computational tools to perform neuroimaging-genetics association studies at a large scale. We will implement and assess on real data the use of novel statistical methodologies and run the statistical analysis on various architectures (grids, clouds), in a unified environment.
Project supported by a Digiteo grant in collaboration with Inria’s KerData Team, MSR-INRIA joint centre, Supélec Engineer School, Imagen project and CEA/Neurospin.
See also:
This project is the following of the A-Brain Project.
Publications
Electronic Journals
-
Gabriel Antoniu, Alexandru Costan, Benoit Da Mota, Bertrand Thirion, Radu Tudoran. A-Brain: Using the Cloud to Understand the Impact of Genetic Variability on the Brain. ERCIM News, April 2012. (on line)
Conferences and workshops
-
Benoit Da Mota, Vincent Frouin, Edouard Duchesnay, Soizic Laguitton, Gaël Varoquaux, Jean-Baptiste Poline, Bertrand Thirion. A fast computational framework for genome-wide association studies with neuroimaging data. 20th International Conference on Computational Statistics (COMPSTAT 2012), Aug 2012, Lamissol, Cyprus. (PDF)
-
Benoit Da Mota, Michael Eickenberg, Soizic Laguittton, Vincent Frouin, Gaël Varoquaux, Jean-Baptiste Poline, Bertrand Thirion. A MapReduce Approach for Ridge Regression in Neuroimaging-Genetic Studies. Data- and Compute-Intensive Clinical and Translational Imaging Applications Workshop (DCICTIA-MICCAI’12), held in conjunction with the 15th International Conference on Medical Image Computing and Computer Assisted Intervention, Oct 2012, Nice, France, To appear. (PDF)
-
Radu Tudoran, Alexandru Costan, Benoit Da Mota, Gabriel Antoniu, Bertrand Thirion. A-Brain: Using the Cloud to Understand the Impact of Genetic Variability on the Brain. 2012 Cloud Futures Workshop, Berkeley, May 2012. To appear.
Posters
-
Benoit Da Mota, Edouard Duchesnay, Vincent Frouin, Gaël Varoquaux, Jean-Baptiste Poline, Bertrand Thirion. Fast vGWAS with Correction for Multiple Tests using Map-Reduce in Cloud or HPC. 18th Annual Meeting of the Organization for Human Brain Mapping, 2012.
-
Benoit Da Mota, Vincent Frouin, Edouard Duchesnay, Soizic Laguitton, Gaël Varoquaux, Jean-Baptiste Poline, Bertrand Thirion. A fast computational framework for genome-wide association studies with neuroimaging data. 5th European meeting on Python in Science (EuroScipy 2012), Bruxelles, Belgium, 2012. (on line)
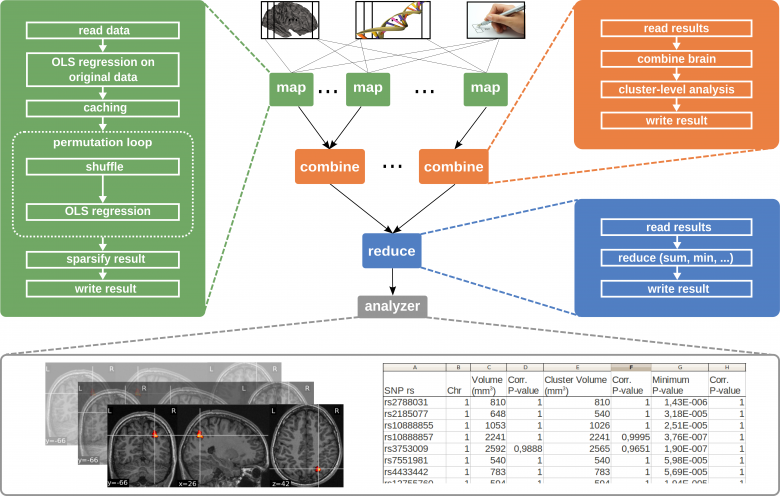
Setting of the MapReduce framework for the Mass-Univariate Linear Model
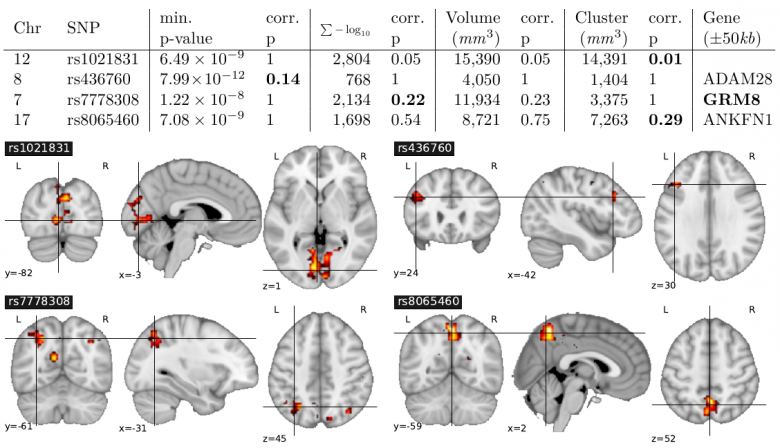
Results of an analysis on IMAGEN dataset with the differents methods and corresponding brain localizations
For details see this paper

