 The objective of Computational Anatomy (CA) is the modeling and analysis of biological variability of the human anatomy. Typical applications cover the simulation of average anatomies and normal variations, the discovery of structural differences between healthy and diseased populations, and the detection and classification of pathologies from structural anomalies. Studying the variability of biological shapes is an old problem (cf. the remarkable book “On Shape and Growth” by D’Arcy Thompson). Significant efforts have been made since that time to develop a theory for statistical shape analysis. Despite all these efforts, there is a number of challenging mathematical issues which remain largely unsolved in general. A particular issue is the computation of statistics on manifolds which can be of infinite dimension (e.g the group of diffeomorphisms).
The objective of Computational Anatomy (CA) is the modeling and analysis of biological variability of the human anatomy. Typical applications cover the simulation of average anatomies and normal variations, the discovery of structural differences between healthy and diseased populations, and the detection and classification of pathologies from structural anomalies. Studying the variability of biological shapes is an old problem (cf. the remarkable book “On Shape and Growth” by D’Arcy Thompson). Significant efforts have been made since that time to develop a theory for statistical shape analysis. Despite all these efforts, there is a number of challenging mathematical issues which remain largely unsolved in general. A particular issue is the computation of statistics on manifolds which can be of infinite dimension (e.g the group of diffeomorphisms).
- There is a classical stratification of the problems into the following 3 levels: construction from medical images of anatomical manifolds of points, curves, surfaces and volumes;
- assignment of a point to point correspondence between these manifolds using a specified class of transformations (e.g. rigid, affine, diffeomorphism);
- generation of probability laws of anatomical variation from these correspondences.
Contacts: Xavier Pennec
The Kernel Bundle framework: Sparse Multiscale Diffeomorphic Deformations
Participants: Stefan Sommer [DIKU], Mads Nielsen [DIKU], François Lauze [DIKU], Xavier Pennec [Correspondant].
This work is performed in collaboration between DIKU (University of Copenhagen) and Asclepios (Inria). It was initiated during a 5 month visit of Stefan Sommer in 2011.
Registration, Diffeomorphic Deformations, Statistical Modeling
In the analysis and modeling of anatomical deformations, we expect deformations to have both large and small scale components. However, we expect these large and small scale deformation to occur at different places. Thus, one would like to represent anatomical deformations with a small number of deformation atoms are different scales that are sparsely distributed across space and scale.
• we propose a multi-scale kernel bundle framework (LDDKBM) that extends the LDDMM framework by incorporating multiple kernels at multiple scales in the registration. Experiments show that the method automatically adapts to the right scales, and it therefore removes the need for classical scale selection methods.
• we derive the Kernel Bundle EPDiff evolution equations, which provide optimal warps in this new framework.
 Longitudinal modeling of the structural changes of the brain in Alzheimer’s disease.
Longitudinal modeling of the structural changes of the brain in Alzheimer’s disease.
Participants: Marco Lorenzi [Correspondant], Xavier Pennec, Giovanni Frisoni [IRCCS Fatebenefratelli Brescia, Italy], Nicholas Ayache.
This work is done in collaboration with LENITEM, IRCCS San Giovanni di Dio Fatebenefratelli, Brescia, Italy.
Alzheimer’s disease, longitudinal model, deformations algorithm, parallel transport, demons algorithm, statis- tical modelling.
This work addresses the analysis and the quantification of the longitudinal structural changes of the brain affected by Alzheimer’s disease (AD). We propose a framework based on the non-rigid registration of brain MRIs using stationnary velocity fields. In 2011, the main scientific developments were:
• Unbiased detection of the structural changes in the brain. The method robustifies the Demons diffeomorphic registration by estimating and removing the multiplicative and additive intensity biases affecting the images.
• Definition of a population-based atlas for the longitudinal brain structural changes. In this work we proposed the Schild’s Ladder as a general method for parallel transporting the subject-specific longitudinal deformation trajectories in a reference space. The work was awarded with the runner-up prize at the “Information Processing in Medical Imaging (IPMI)” conference in Irsee, Germany. Other transport methods from the Lie group theory have been successively proposed and investigated [62].
• Group-wise statistical analysis of the brain longitudinal changes in multiple time points. The framework has been applied for the analysis of the longitudinal brain changes in healthy subjects at risk of AD, and the results showed an accelerated progression of atrophy for the subjects positive to the marker of Alzheimer Aβ42.
Finally, the above framework has been promoted to the neuroscience community as diagnostic tool and support for clinical trials.
 Statistical Analysis of White Matter Fiber Bundles
Statistical Analysis of White Matter Fiber Bundles
Participants: Stanley Durrleman [Correspondant], Pierre Fillard [Parietal, INRIA Saclay], Xavier Pennec, Alain Trouvé [CMLA, ENS Cachan], Nicholas Ayache.
Computational anatomy, Fiber bundle, Fiber tract, Diffusion imaging, Diffeomorphic registration, Atlas construction, Variability analysis, Currents
This work is an application of the generic morphometric method developed in 2009 to the statistical analysis of the shape and texture of white matter fiber bundles extracted form Diffusion Tensor Images (DTI).
• Registration, Atlas Estimation and Variability Analysis of White Matter Fiber Bundles Modeled as Currents.
 Comparison of endocranial ontogenies in chimpanzees and bonobos
Comparison of endocranial ontogenies in chimpanzees and bonobos
Participants: Stanley Durrleman [Correspondant], Xavier Pennec, Alain Trouvé [CMLA, ENS Cachan], Nicholas Ayache, José Braga [AMIS, Univ. Toulouse 3].
This work has been performed in the context of the INRIA collaborative project ARC 3D-Morphine (PI: Sylvain Prima, IRISA) and a follow-up collaboration with José Braga at Université Paul Sabatier, Toulouse.
Endocranium, Virtual anthropology, Pan, Ontogeny, Shape regression, Spatiotemporal registration
 Statistical Modelling of Cardiac Growth and Deformation from Medical Images
Statistical Modelling of Cardiac Growth and Deformation from Medical Images
Participants: Kristin McLeod [Correspondant], Tommaso Mansi, Adityo Prakosa, Maxime Sermesant, Xavier Pennec.
Parts of this work were performed within the framework of the EU project Care4me ITEA2, and the INRIA ARC Sirap, in collaboration with St Thomas Hospital, King’s College London, the REO team from INRIA Rocquencourt and Necker Paediatric Hospital in Paris.
Heart, statistical modelling, registration, Demons algorithm, LDDMM algorithm, currents, motion tracking,
This work builds on the statistical analysis framework for surfaces developed by Durrleman and Mansi in 2009.
• The iLogDemons motion tracking algorithm of Mansi et. al was applied to a data-set of 15 subjects and 1 phantom each with a cine-MR, tagged-MR and echocardiography sequence as a part of the STACOM workshop challenge at MICCAI 2011. The paper received the Best Paper Award for the Motion Challenge.
• The work of Mansi et. al for a statistical model of cardiac growth in the right ventricle was extended to the left ventricle to obtain a bi-ventricular cardiac growth model for patients with repaired tetralogy of Fallot.
• The preliminary analysis of a statistical model for reduced blood flow simulations in the pulmonary artery proposed in 2010 is currently being extended to a journal version to further analyse the method on a larger data-set.
 Statistical Modeling of Shapes Using Trees of Locally Affine Transformations
Statistical Modeling of Shapes Using Trees of Locally Affine Transformations
Participants: Christof Seiler [Correspondant], Xavier Pennec, Mauricio Reyes [Institute for Surgical Tech- nology and Biomechanics, University of Bern, Switzerland].
This work is performed in the context of the joint PhD of Christof Seiler at the Institute for Surgical Technology and Biomechanics, University of Bern, Switzerland and Asclepios INRIA.
Registration, Parametrization of Diffeomorphic Deformations, Trees of Transformations, Statistical Modeling, Mandibles, Femurs
The goal of this work is to analyze anatomical shapes through deformations defined with few but important and intelligible parameters. Advances towards this goal were the following in 2011.
• Fusion of the Log-demons registration and the Log-Euclidean polyaffine framework. The results of the new registration method applied to femur CT’s was presented at SPIE.
• Decomposition of diffeomorphic deformations into a tree of locally affine transformations applied to mandible CT’s. This work won the young scientist award at MICCAI 2011 in Toronto.
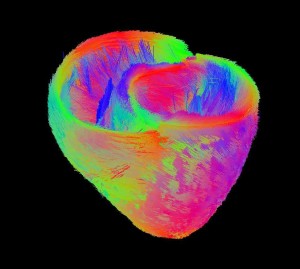
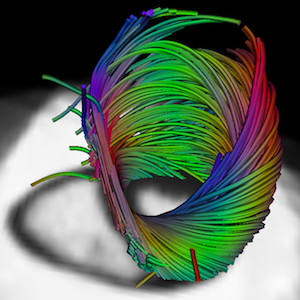 Atlas of Cardiac Fiber Architecture from DT-MRI
Atlas of Cardiac Fiber Architecture from DT-MRI
Participants: Hervé Lombaert, Hervé Delingette [Correspondant], Nicholas Ayache, Jean-Marc Peyrat, Pierre Croisille.
This work is a collaboration between Creatis, Lyon and INRIA Sophia Antipolis, including members of Ecole Polytechnique of Montreal. Financial support is partly from the National Science and Engineering Research Council of Canada, and the Michael Smith Foreign Study Program.
atlas,registration,segmentation
The variability of the cardiac fiber architecture has been investigated in a human population. An automatic method has been developped to construct an atlas of DTMRI images. A statistical analysis has been carried on using the Log-Euclidean metric.
• An automatic method has been developped to construct an atlas of DTMRI images. The first human
atlas of DTMRI has been build. This work received the Best Paper Award at the FIMH 2011 conference in New York City.
• Results have also been published where the variability of the cardiac architecture is measured using the Log-Euclidean metric.
• The variability of the cardiac laminar sheets in human DTMRI has been studied.
• Differences in a population of normal and abnormal hearts have been investigated.



 Non invasive cardiac personalisation
Non invasive cardiac personalisation
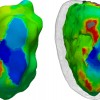 Simulation of ventricular tachycardia re-entry circuit
Simulation of ventricular tachycardia re-entry circuit
 heartMeshFine
heartMeshFine
 Electrophysiology
Electrophysiology
 Cardiac Fibres from in vivo Diffusion Tensor Imaging
Cardiac Fibres from in vivo Diffusion Tensor Imaging
